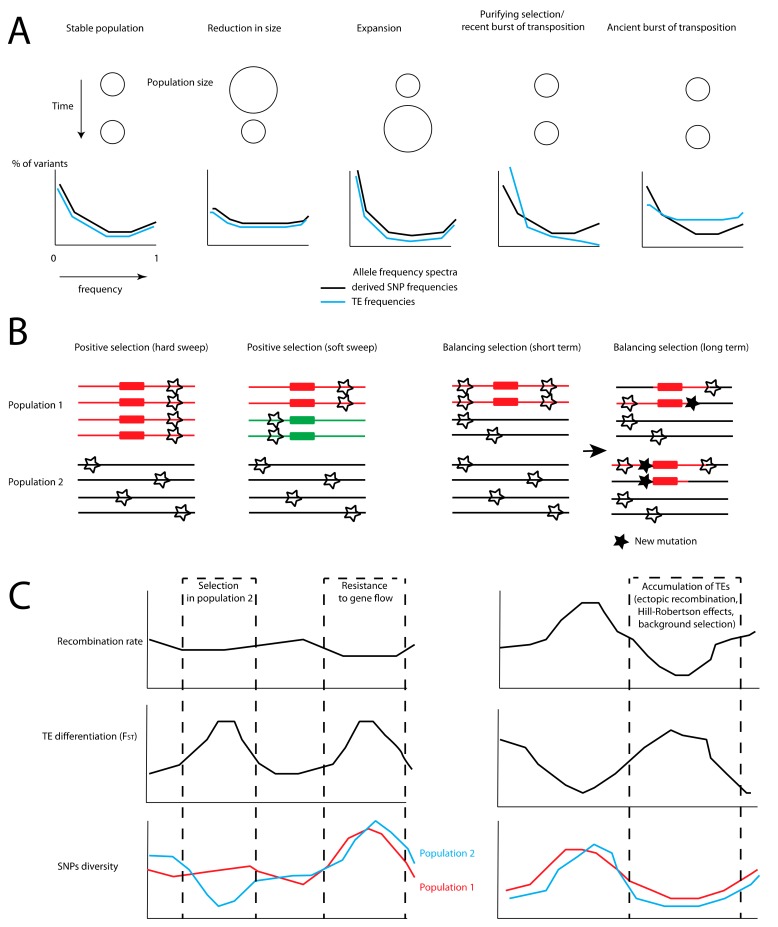
Figure 1.
Summary of mechanisms impacting the diversity and frequency of transposable elements (TEs), and their impact on flanking sequences. (A) Demographic changes affect the frequency spectra of both TEs and single nucleotide polymorphisms (SNPs) in a similar way, assuming neutrality and a constant rate of transposition. Reductions in effective population sizes should lead to an excess of alleles at intermediate frequencies, while population expansions may lead to an excess of singletons. On the other hand, purifying selection on TEs should lead to an excess of singletons compared to SNPs. Variable rates of transposition may also lead to discrepancies in the spectra between SNPs and TEs. (B) TEs involved in adaptation may be detected through their changes in frequencies, but also through the signature left in flanking regions. In the case of positive selection, longer, younger haplotypes should be found nearby positively selected insertions. The similarity of selected haplotypes may be very high in the case of a recent hard sweep, where the insertion is immediately selected and rises in frequency. It may be lower in the case of a so-called soft sweep, where selection either acts after the insertion has already reached an appreciable frequency in the population, or when two insertions with a similar effect on fitness appear at the same time. Positive selection should also result in higher differentiation at the selected locus compared to populations where selection is not acting. On the other hand, balancing selection may lead to signatures of partial selective sweep when it is recent. Since the selected alleles may be maintained through long periods of time, they have more time to recombine and accumulate new mutations than neutral haplotypes, leading to a narrow signature of high diversity. Since alleles under balancing selection tend to introgress into new populations, and have high diversity, low differentiation is expected at these sites. (C) Left panel: Given a constant recombination rate, positive and linked selection in a given population (here, a population of two) may increase differentiation and reduce diversity at selected TEs and flanking regions compared to the rest of the genome. On the other hand, if TEs play a role in incompatibilities after secondary contact, a signature of both elevated differentiation and diversity may be expected. Right panel: However, an excess of TEs in regions of reduced polymorphism, higher differentiation, and lower recombination may be caused by different mechanisms such as purifying selection. This can be due to a reduced effective rate of transposition in regions of high recombination due to deleterious ectopic exchanges, and/or because of the larger-scale effect of selection that accelerates lineage sorting and the differentiation of TEs in regions of low recombination.