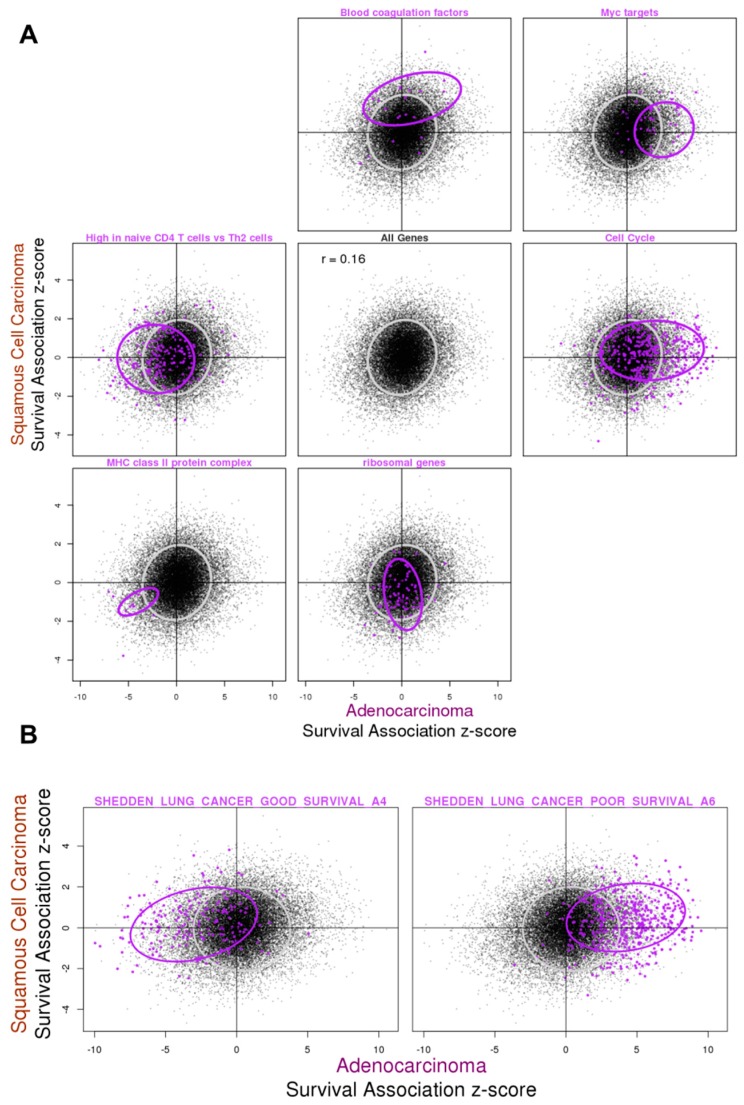
Figure 3.
Gene expression survival associations in ADC and SQCC with selected gene sets highlighted. Summary z-scores from the meta-analysis of gene expression survival association based on the Cox-PH model were plotted for all genes, with the results from ADC on the x-axis and results from SQCC on the y-axis. The Pearson correlation coefficient for all genes was 0.16. (A) Genes from the selected gene sets coming from the MSigDB gene set collections were highlighted. A selection of gene sets that showed the different patterns for ADC and SQCC association were also highlighted, including: “Blood coagulation factors” from MODULE_131 in the c4 computational gene set collection, “Myc targets” from HALLMARK_MYC_TARGETS_V2 in the hallmark gene set collection, “High in Naïve CD4 T cells vs. Th2 cells” from GSE22886_NAIVE_CD4_TCELL_VS_12H_ACT_TH2_UP in the c7 immunologic signatures gene sets collection, “Cell Cycle” from REACTOME_CELL_CYCLE in c2.cp curated the canonical pathway gene set collection, “MHC class II protein complex components” from GO_MHC_CLASS_II_PROTEIN_COMPLEX in c5.cc GO cellular component gene set collection, and “ribosomal genes” from KEGG_RIBOSOME in the c2.cp curated canonical pathway gene set collection. (B) Two prognosis-associated gene sets summarized from a previous lung adenocarcinoma study, SHEDDEN_LUNG_CANCER_GOOD_SURVIVAL_A4 and SHEDDEN_LUNG_CANCER_POOR_SURVIVAL_A6 in the c2.cgp curated chemical and genetic perturbation gene set collection, were also highlighted. Ellipsoidal boundary wraps were around the 75% highest density/minimum volume for all genes (gray) or selected genes (purple).