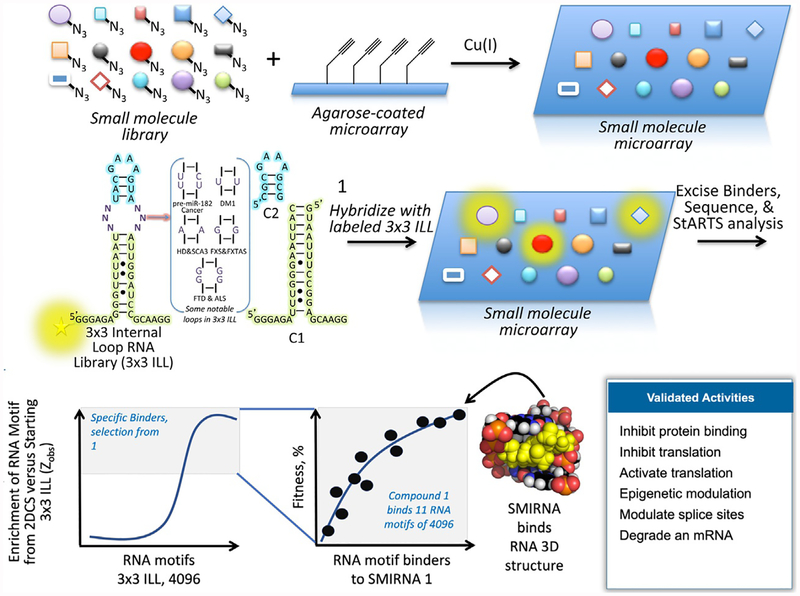
Fig. 1.
Discovering the three-dimensional RNA folds that bind small molecules using a library-vs.-library screen named 2-Dimensional Combinatorial Screening (2DCS). A small molecule library is site-selectively immobilized (shown) or absorbed onto agarose-coated microarrays. The arrays are incubated with a labeled RNA fold library that displays discrete structural elements (such as the 3 × 3 internal loop library (ILL)) in the presence of competitor oligonucleotides that mimic regions common to all library members (C1 and C2, for example). Bound RNAs are excised from the surface, and their identities are determined by RNA-seq. Statistical analysis of RNA-seq data reveals privileged RNA fold-small molecule interactions that inform design of SMIRNAs that modulate RNA biology. Along with 2DCS, we developed an approach named Structure-Activity Relationship Through Sequencing (StARTS), which allows for facile annotation of 2DCS data by scoring the relative affinity and selectivity of RNA motifs that bind to a small molecule (a metric of small molecule “fitness”).