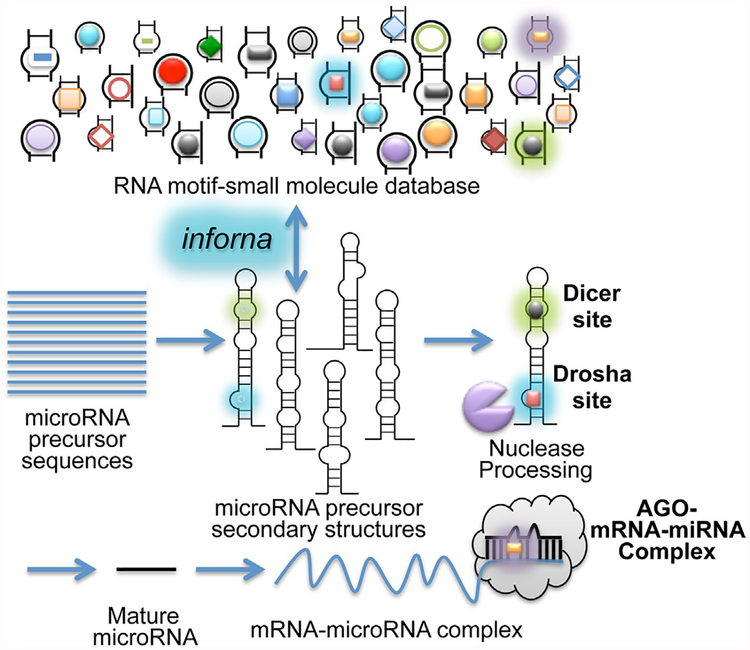
Fig. 2.
Design of SMIRNA using Inforna, a lead identification strategy for RNA targets. Privileged RNA fold-small molecule interactions identified by 2DCS are annotated and deposited into a database. Inforna designs lead small molecules by comparing the folds in the database to folds in a cellular RNA target. Illustrated is implementation of Inforna to identify SMIRNAs for miRNA targets.