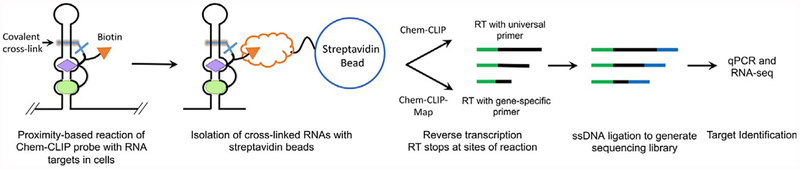
Fig. 3.
Overview of the target validation and profiling method Chemical Cross-Linking and Isolation by Pull-down (Chem-CLIP). A Chem-CLIP probe is an SMIRNA appended with a cross-linking module (typically the nitrogen mustard chlorambucil) and a biotin purification module. Thus, RNA targets that bind the SMIRNA are brought into close proximity with the cross-linking module, forming a covalent adduct upon reaction. In brief, an appropriate cell line is incubated with the Chem-CLIP probe, followed by isolation of total RNA. RNAs that reacted with the Chem-CLIP probe are isolated and purified via its biotin tag and streptavidin beads. After elution of the adducts, the RNAs are reverse transcribed with either a universal primer (Chem-CLIP to identify all targets) or a gene-specific primer (Chem-CLIP-Map to identify the SMIRNA binding site). After RT, a single stranded DNA adaptor is ligated to enable generation of a sequencing library; qPCR and sequencing (RNA-seq) are then completed.