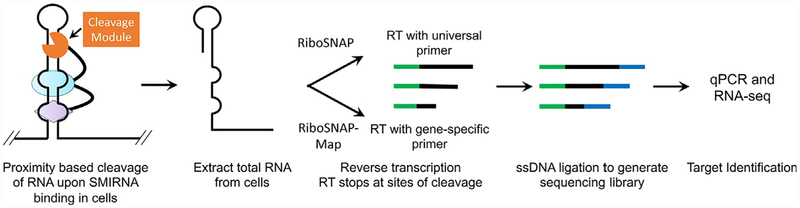
Fig. 4.
Overview of the target validation and profiling method Small Molecule Nucleic Acid Profiling by Cleavage Applied to RNA (RiboSNAP). A RiboSNAP probe is an SMIRNA appended with a cleavage module, which can be activated by light (such as N-HPT), recruit a nuclease to effect cleavage at the target site (RIBOTACs), or cleave the target directly (bleomycin). RNA targets that bind the SMIRNA bring the cleavage module into close proximity such that the RNA is cleaved. Briefly, cells are treated with a RiboSNAP probe and total RNA is isolated. In contrast to Chem-CLIP studies, total RNA can be analyzed directly, avoiding the pull-down step. Reverse transcription is either completed with a universal primer to identify all targets (RiboSNAP) or with a gene-specific primer to localize the SMIRNA binding site (RiboSNAP-Map). A sequencing library is then generated by ligation of a single stranded DNA adaptor and subsequent qPCR and RNA-seq analysis.