SUMMARY
Although animals have evolved multiple mechanisms to suppress transposons, “leaky” mobilizations that cause mutations and diseases still occur. This suggests that transposons employ specific tactics to accomplish robust propagation. By directly tracking mobilization, we show that, during a short and specific time window of oogenesis, retrotransposons achieve massive amplification via a cell-type-specific targeting strategy. Retrotransposons rarely mobilize in undifferentiated germline stem cells. However, as oogenesis proceeds, they utilize supporting nurse cells, which are highly polyploid and eventually undergo apoptosis, as factories to massively manufacture invading-products. Moreover, retrotransposons rarely integrate into nurse cells themselves but, instead, via microtubule-mediated transport, preferentially target the DNA of the interconnected oocytes. Blocking microtubule-dependent intercellular transport from nurse cells significantly alleviates damage to the oocyte genome. Our data reveal that parasitic genomic elements can efficiently hijack a host developmental process to propagate robustly, thereby driving evolutionary change and causing disease.
INTRODUCTION
As the most abundant residents in the genomes of nearly all eukaryotes, transposons represent a potential source of genome instability (Chuong et al., 2017; Kazazian and Moran, 2017). Although the hosts have evolved multiple mechanisms to suppress transposable elements, “leaky” mobilizations that cause mutations and diseases still occur (Chuong et al., 2017; Kazazian and Moran, 2017; Weick and Miska, 2014; Yang et al., 2017). For example, Doc element transposed into the white locus of Drosophila genome, which allowed Morgan to identify the first documented white-eye fly and to lay the basis of modern genetics (Driver et al., 1989; Morgan, 1910). Other classic examples are LINE1 mobilizing into the genomic locus of FVIII or APC led to hemophilia or colon cancer, respectively (Dombroski et al., 1991; Miki et al., 1992). These findings suggest that transposons potentially employ developmental regulation to accomplish robust propagation, but the underlying mechanism remains elusive.
In animal gonads, it has been proposed that PIWI-interacting RNAs (piRNAs) suppress transposons to ensure the faithful transmission of genetic information from one generation to the next (Aravin et al., 2006; Girard et al., 2006; Grivna et al., 2006; Ishizu et al., 2012; Saito et al., 2006; Siomi et al., 2011; Vagin et al., 2006; Weick and Miska, 2014). The piRNA binding partners–the PIWI clade Argonaute proteins (Ago3, Aub, and Piwi in Drosophila)–lie at the center of the pathway for piRNA biogenesis and function (Brennecke et al., 2007; Li et al., 2009). Reciprocal cycles of piRNA-guided RNA cleavage, which are catalyzed by Aub and Ago3, expand the piRNA population while simultaneously destroying transposon transcripts (Brennecke et al., 2007; Gunawardane et al., 2007). Meanwhile, Ago3 cleavage also initiates phased piRNA production, which guides Piwi for silencing of transposon transcription in the nucleus (Han et al., 2015; Mohn et al., 2015; Senti et al., 2015; Wang et al., 2015). Given the fact that piRNA pathway perturbation unleashes these parasitic elements from silencing, it provides a unique opportunity to study the underlying mechanisms by which transposons exploit gametogenesis regulation to enhance their mobility. However, in most studies, transposon activity is typically assessed by the level of their mRNAs from whole tissues (Klattenhoff et al., 2009; Li et al., 2009; Zhang et al., 2011). Little is known about transposon activity at the transposition level: whether transposons indeed mobilize; if so, at which developmental stages and in which cell types do transpositions occur.
In this study, we developed approaches to track transposon mobilization with spatiotemporal resolution during Drosophila oogenesis, which is a well-characterized process and has served as a critical model system to study the function of piRNA pathway (Mahajan-Miklos and Cooley, 1994; Siomi et al., 2011; Spradling, 1993). As oogenesis proceeds, one germline stem cell gives rise to 15 supporting nurse cells and one oocyte. Although undergoing programmed cell death at the end of oogenesis, during the process of oocyte development, nurse cells produce the vast majority of cytoplasmic constituents/nutrients for oocyte from their highly polyploid genome (Mahajan-Miklos and Cooley, 1994; Spradling, 1993). Here, we show that retrotransposons barely mobilize in germline stem cells. Upon differentiation, they utilize differentiated nurse cells to massively manufacture their invading products, but, seldom transpose into nurse cell DNA. Instead, via microtubule-mediated transport, retrotransposons selectively target the DNA of oocyte, the only ovarian cell that founds the next generation. Our data therefore demonstrate that retrotransposons exploit a hijacking tactic to robustly propagate in the host genome during oogenesis.
RESULTS
An eGFP reporter to monitor the I-element mobilization
Inspired by previous studies that employ a disrupting intron to monitor mobilization of retrotransposons (Boeke et al., 1985; Jensen and Heidmann, 1991; Moran et al., 1996), which represent the majority of transposons in the animal genome (Chuong et al., 2017; Kazazian and Moran, 2017), we constructed a reporter whereby eGFP signal is only produced after retrotransposition (Figure 1A and S1A). In essence, the eGFP reporter, which is placed at the 3′ end of the transposon in the antisense orientation, is disrupted by an intron in the same orientation as the transposon (Figure 1A and S1A). The reporter can only produce eGFP when the transposon transcript is spliced, reverse transcribed, and its cDNA sequence is integrated back into the genome (Figure 1A), therefore serving as an ideal tool to track transposon mobilization with single-cell resolution.
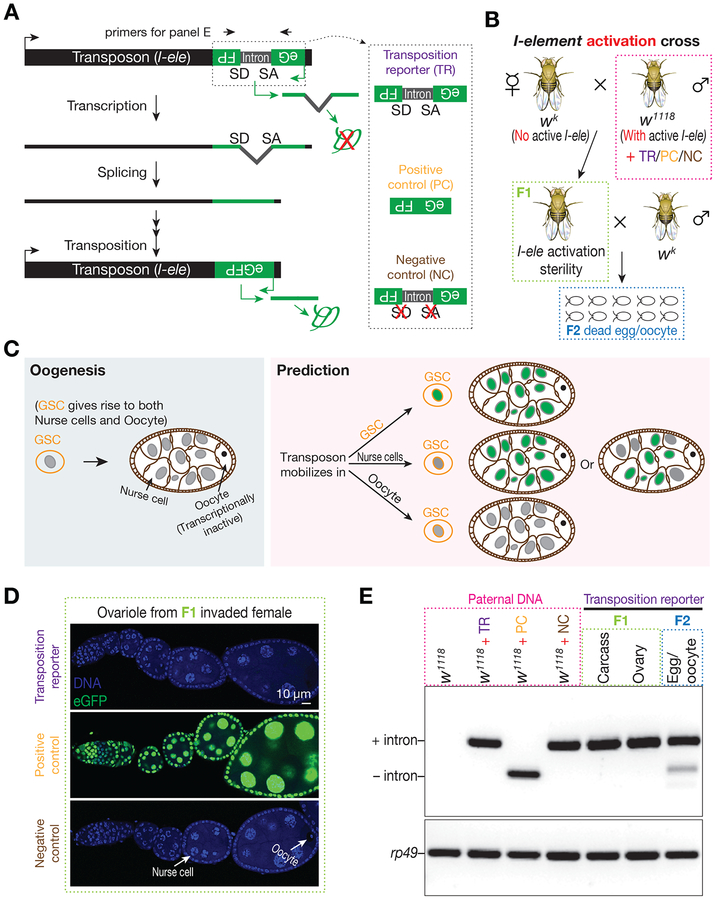
Figure 1. Tracking I-element mobilization during oogenesis with an eGFP reporter reveals oocytes are preferentially targeted for transposition.
(A) Schematic desgin of eGFP reporter to track I-element mobilization. An eGFP reporter is inserted into the 3’ UTR of I-element sequences in the antisense direction. eGFP expression is blocked by an intron, which is in the same orientation of transposon and therefore is unable to be spliced during eGFP transcription. Only transposition can generate a copy of DNA without the dirupting intron, thereby producing eGFP signal. A construct without disrupting intron serves as positive control to test the potential sensitivity of reporter. A construct with mutations on the splicing acceptor and donor sites serves as negative control.
(B) The genetic setup to trigger I-element mobilization. Without protection from the I-element-silencing piRNAs, which are typically maternally deposited, paternally inherited I-elements become active in invaded progeny. These offspring were crossed with wk males to purify the oocytes from their ovaries. The I-element containing fathers carry either transposition reporter or one of the control constructs.
(C) A simplified cartoon to illustrate Drosophila oogenesis and the potential outcomes from reporter.
(D) Detecting eGFP expression in invaded ovaries carrying each construct. Transposition reporter does not give signal from any ovarian cells in I-element activated progeny. The strong signal from positive control construct highlights the potentially high sensitivity of reporter.
(E) Probing transposition events by PCR. Transpositions in oocyte generate intron-removed DNA, which gives rise to a short PCR product. Primers are depicted in panel A. Samples were boxed with different color to match the corresponding animals depicted in panel B.
Because of its similarity to the mammalian LINE1 transposon and the availability of a powerful genetic system to induce robust activation (Brennecke et al., 2008; Bucheton et al., 1984; Picard, 1976; Seleme et al., 2005), we chose the non-LTR retrotransposon, I-element, to test the designed reporter. When male flies carrying the active I-elements mate with females lacking active copies, which could not maternally deposit I-element-silencing piRNAs, I-elements escape from the piRNA pathway-mediated silencing, becoming active in the resulting invaded progeny and leading to animal sterility (Brennecke et al., 2008) (Figure 1B and S1B). To track the mobilization of the I-element in these offspring, we introduced one copy of I-element that contains eGFP reporter into the genome of I-element-containing males (Figure 1B and S1B). To ensure a single copy of eGFP reporter would give an eGFP signal without the disrupting intron, flies carrying an intronless reporter were used as a positive control (Figure 1A). Additionally, a non-spliceable construct, which contains mutated splicing acceptor and donor sites (Figure 1A), serves as a negative control to verify that eGFP production relies on intron removal during transposition.
Based on the reporter design, we predicted that the mobilizations of I-element transgene would mark the cells that harbor these events as eGFP positive (Figure 1C). Surprisingly, in invaded females for which the I-element mobilizations cause sterility, we did not detect eGFP signal from any ovarian cell types (Figure 1D). In contrast, flies harboring a single copy of the intronless construct (positive control) gave robust eGFP signal in all ovarian cell types (Figure 1D), highlighting the potentially high sensitivity our reporter would achieve. These unexpected results indicate that I-element rarely mobilizes into nurse cells and germline stem cells, which would mark all germ cells derived from them as eGFP positive (Figure 1C). We hence proposed that upon activation, I-elements preferentially transpose into the genome of oocytes. The transcriptionally inert character of the oocytes (King and Burnett, 1959; Mahowald, 1972; Mahowald and Tiefert, 1970) would explain why there is no eGFP expression from the newly inserted reporters. Supporting I-elements selectively target oocytes for mobilization, the oocytes show strong enrichment of I-element mRNA and Orf1 proteins (Brennecke et al., 2008; Orsi et al., 2010; Seleme et al., 2005; Van De Bor et al., 2005) (Figure S2).
To directly test the occurrence of I-element mobilization in oocytes, we next purified them by mating invaded females with male flies that do not contain the eGFP reporter (invaded females still lay dead eggs/oocytes upon mating, Figure 1B), and measured the appearance of the intron-removed reporter in their genome by Polymerase Chain Reaction (PCR). We indeed detected a clear intron-removed PCR product from the DNA of oocytes, but not the DNA from ovaries or other somatic cell samples (Figure 1E), indicating that the I-element transgene preferentially inserts into the oocyte genome. We noticed that the intron-removed PCR product has relatively weak intensity, comparing with the product from intron-retained DNA. This may reflect that the splicing efficiency of the intron is not at the optimal level. Comparing with the oocytes from invaded females, the intron-removed PCR product was not detected in the oocytes laid from the control progeny (Figure S1B and S1C). Since these flies carry the same eGFP reporter transgene but are equipped with I-element-silencing piRNAs, this result validated that the intron-removal depends on I-element activation.
Native I-elements also preferentially target the oocyte DNA
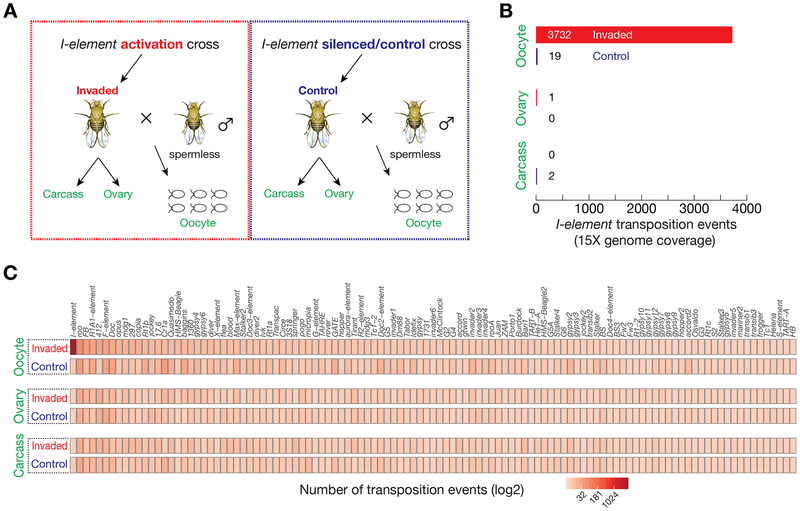
To test whether native I-elements in fly genome also selectively target the oocyte DNA, we employed paired-end genome sequencing and computational analysis to detect transposition events from the oocytes and ovaries of I-element-invaded progeny. We crossed invaded females with spermless males to collect their oocytes that do not contain the DNA from sperm (Figure 2A and S3A). Meanwhile, we also extracted the DNA from the gonad-ectomised carcasses to examine whether native I-elements mobilize in somatic cells (Figure 2A and S3A). To estimate the false-positive mobilization rate for each sample type, we additionally sequenced the DNA of oocytes, ovaries, and ovary-ectomised carcasses from control progeny, which have identical genotype as invaded offspring but with I-elements being silenced by the piRNA pathway (Figure 2A and S3A). With 15× genome coverage, we only detected < 5 potential I-element insertion events from ovary or ovary-ectomised carcass, irrespective of invaded or control progeny being examined (Figure 2B, 2C, and S3B). This suggests that endogenous I-elements barely mobilize in ovary and other somatic cells. In stark contrast, the oocytes from invaded progeny possessed 3732 new I-element insertions (Figure 2B, 2C, and S3B), > 196 times greater than predicted by the false-positive rate, which is estimated from the oocytes laid by controls. These findings, in accordance with the results from our eGFP reporter, strongly support the conclusion that I-elements, avoid nurse cells and other somatic cells for integration, but instead, selectively target the oocyte genome.
Figure 2. Native I-elements selectively target oocyte genome for propagation.
(A) Samples used (marked as green) for genome sequencing to detect new transposon insertions. A detailed cross-scheme is presented in Figure S3.
(B) Transposon insertion events from I-elements.
(C) Transposition events from all transposons.
For the genetic setup that used to activate I-elements, it might also stimulate the activation of other transposon families in fly genome, therefore contributing to sterility. Our data from whole-genome sequencing enable us to examine this possibility. Except for I-element, all other transposon families possessed indistinguishable mobilization rate between invaded and control progeny (Figure 2C and S3B), regardless of the DNA being extracted from oocytes, ovaries, or gonad-ectomised carcasses. These data indicate that our genetic system only induces the mobilization of I-element, and that it is the propagation of I-elements that causes sterility of the invaded offspring.
A group of transposons selectively mobilizes into oocyte DNA
Do other retrotransposons have the same cell type preference for targeting as I-element? To stimulate transposon activity for measurement, we next depleted the core piRNA pathway components–Ago3 and Aub–by RNAi in fly ovaries and then globally measured transposon mobilization events in ovaries, oocytes, and ovary-ectomised carcasses (Figure S4). Serving as controls, we depleted the non-essential White proteins by using the same strategy to generate the genetically identical progeny (except the shRNA constructs for RNAi, Figure S4). These control animals were used to define the number of potential new insertions when the piRNA pathway is intact. Similarly, spermless males were used to obtain oocyte DNA (Figure S4).
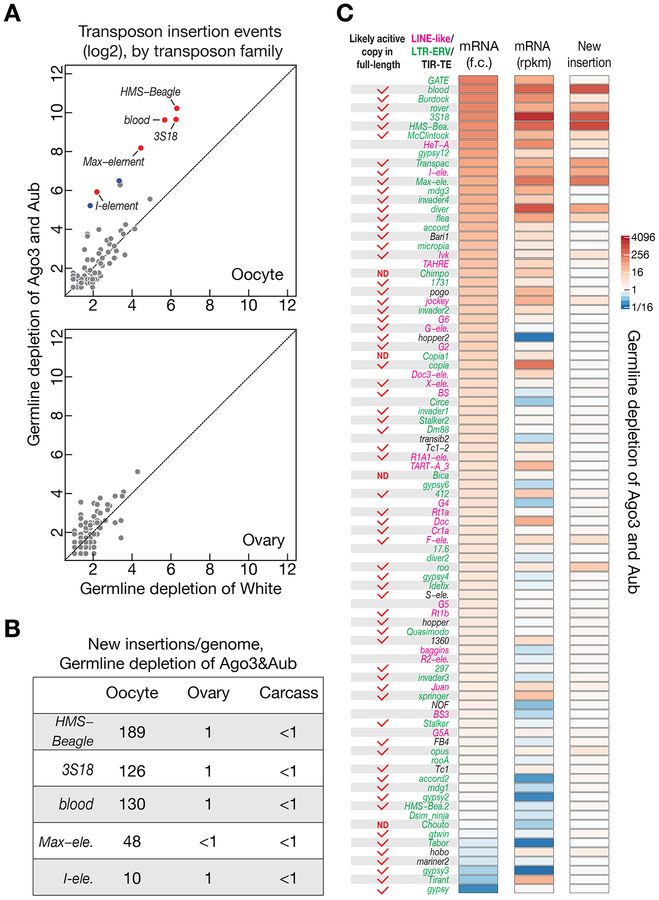
When plotting the potential new transposon insertions that were detected for each transposon family from the ovary or carcass genome, Ago3&Aub-depleted flies had almost identical profiles as White-depleted progeny (Figure 3A and S5A). This suggests that transpositions rarely happen in these tissues irrespective of perturbation of the piRNA pathway. In contrast, immense transpositions occurred in oocytes upon blocking the piRNA pathway, but not in control oocytes (Figure 3A and S5B). Five transposon families showed more than a 10-fold significant increase (HMS-Beagle, 3S18, blood, Max-element, and I-element), and 2 families significantly increased by more than 5-fold, but less than 10-fold (Transpac and diver). flea is the only transposon that made less than 5-fold increase, but still passed the significance test (p-value of the T-test: 0.03, 4-fold increase). Aside from I-element, the rest of the active transposons all belong to the LTR (Long Terminal Repeat) proviral family, indicating that these proviruses, although were endogenized by Drosophila melanogaster a long time ago, still maintain high capability for mobilization.
Figure 3. A group of retrotransposons preferentially mobilizes into oocyte genome.
(A) Genome sequencing to globally probe new transposition events in White (control) or Ago3&Aub depleted progeny. Upon piRNA pathway perturbation, retrotransposons preferentially insert into oocyte genome. Each dot represents one transposon family. Red: significantly changed for more than 10-fold. Blue: significantly changed for more than 5-fold but less than 10-fold.
(B) The number of new insertions (per genome) detected for the top 5 active transposons in Ago3&Aub depleted progeny. We found that the number of new insertions shows linear correlation with the number of genome coverage (Figure S5C).
(C) Upon piRNA perturbation, neither fold-change nor the absolute mRNA abundance in ovaries could faithfully reflect transposition activity in oocytes. RNA-Seq data and genome sequencing data were generated from the flies have identical genotypes. The change of mRNA abundance was quantified as fold change, and transposition events were calculated by subtracting the potential false positive numbers (from White depletion) from the insertion numbers detected in the oocytes laid by Ago3&Aub depleted progeny. The insertion events detected from 5× genome coverage were used to generate figures for panel A and C. ND = not determined yet.
Notably, the top 4 active transposons (HMS-Beagle, 3S18, blood, and Max-element) made 91% of total insertions detected in Ago3&Aub depleted oocytes. With 5.4× genome coverage, we identified in total 3062 new insertions from all transposon families analyzed, and 2799 of them were from these four most active families (1092 from HMS-Beagle, 713 from 3S18, 725 from Blood, and 269 from Max-element). To precisely calculate the number of new insertions per genome for each of them, we first tested whether the number of new integrations detected by our approach correlates with the fold of genome coverage. By randomly sampling the sequencing reads to achieve 5-fold, 10-fold, 20-fold, 30-fold, 40-fold, and 50-fold coverage of the Drosophila genome, we found that the number of insertion events obtained from oocyte shows linear correlation with the number of genome coverage (R2 = 0.99 for all 4 families, Figure S5C). Correspondingly, we predicted that there are 189 new insertions per genome for HMS-Beagle, 126 new integrations for 3S18, 130 for blood, and 48 for Max-element (Figure 3B and S5C). Meanwhile, the number of potential new insertions detected from ovary or carcass was negligible, highlighting the low false-positive rate of our approach (Figure 3B and S5C).
LTR (Proviral) and LINE-type transposons have distinct landing preferences
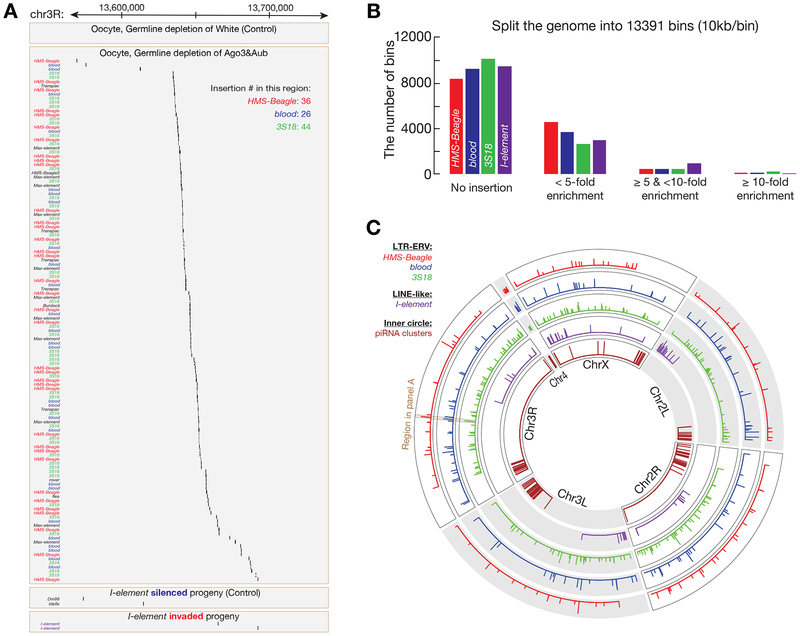
Visualization of insertion sites from browser suggested a non-random distribution of integration sites across the genome (Figure 4A). We thus tested whether retrotransposons exhibit an integration preference across the whole genome. For this purpose, we used all of the sequencing reads from oocyte samples and analyzed the landing “hotspots” for the four transposons that had > 5000 new insertions in our study: HMS-Beagle, 9199 insertions; blood, 6356; 3S18, 5977; I-element, 5071. Splitting the genome into 13391 bins of 10kb width, 62–75% of bins did not have any detected insertions (Figure 4B). For the three LTR retrotransposons, 4–5% of bins had the number of insertions 5-fold greater than the number of insertions would expected if there was no integration preference (Figure 4B); ~1% of bins showed more than 10-fold greater (Figure 4B). For the LINE type I-element, 7.5% of the bins had greater than 5-fold the number of insertions expected, and 0.5% of bins contained more than 10-fold enrichment insertions (Figure 4B). To keep our analysis stringent, we define hotspots as genomic regions that have at least 10-fold enrichment: 103 hotspots for HMS-Beagle, 116 for blood, 219 for 3S18, and 63 for I-element (Figure 4B, 4C, and Table S1).
Figure 4. LTR-ERV and LINE-type transposons show distinct landing preferences.
(A) One genomic region that is targeted by LTR transposons, but not I-element, from Chromosome 3R is shown.
(B) Hotspot analysis for the retrotransposons that made more than 5000 new insertions in our study. The fly genome is split into 13391 bins. Assuming there is no landing preference, the number of theoretical insertions per bin is calculated. And this number is used to define the enrichment for each bin (based on the insertions practically detected). The regions that have ⩾ 10-fold enrichment are classified as hotspots.
(C) Circos plot to display the hotspots for all four retrotransposons. Inner circle shows piRNA clusters in fly genome. LTR-ERV transposons share significant number of hotspots, but not with the LINE-type I-element.
Visualizing new landing sites showed that LTR retrotransposons preferentially target the same genomic loci, i.e., share significant number of hotspots (Figure 4A and 4C). Notably, these loci are distinct from the LINE type I-element. For example, for the 103 hotspots from HMS-Beagle, 42 were shared with blood and 49 overlapped with 3S18, but only 3 were shared with I-element (Figure 4A and 4C). Similarly, blood shared 57 hotspots with 3S18, but only 4 with I-element (Figure 4A and 4C). In total, there were 31 genomic loci that are preferentially targeted by all of the three proviral transposons (Figure 4A and 4C). These data indicate that, distinct from I-element, LTR transposons select target sites for integration via a common mechanism. Perhaps their integrases are recruited to the oocyte genome through the same host factor, resembling the function of p75/LEDGF for HIV integration (Cherepanov et al., 2004).
The fly genome contains 142 piRNA clusters, which are enriched with fragmented transposons, to serve as source for piRNA production in Drosophila ovary (Brennecke et al., 2007). It is unclear what leads to the accumulation of transposon remnants in these loci. Perhaps transposons preferentially integrate into these cluster regions. Alternatively, transposons may not favor these clusters for integration, but rather the enrichment reflects the final outcome of selection through evolution, i.e., when animals confront deleterious transposon activation, randomly landing into clusters propels piRNA production, thus improving the chance for survival. Our data favor the latter possibility (Figure 4C). For the three LTR transposons, we only detected 4 (HMS-Beagle and 3S18) or 5 (blood) hotspots, from hundreds of them, overlapped with piRNA clusters (Figure 4C). Additionally, none of the hotspots from I-element overlapped with these clusters (Figure 4C). Since the cluster sites comprise 3.5% of fly genome (Brennecke et al., 2007), our data suggest that these retrotransposons do not preferentially mobilize into piRNA clusters. However, we cannot exclude the possibility that the repetitive feature of clusters leads to an underestimation of integration events in these regions.
Transposon mobilization appears to cause DNA damage in developing oocytes
Retrotransposons initiate integration by creating DNA breaks (Feng et al., 1996), which can be visualized by staining with a marker of DNA break, a phosphorylated version of histone variant (γ-H2Av). Although mutating the piRNA pathway appears to enhance the DNA damage generated from meiotic recombination (Klattenhoff et al., 2007), whether the mobilizations of retrotransposons lead to the accumulation of DNA breaks in later developing oocytes is still unclear. We next measured the signal of γ-H2Av in developing oocytes from the stage 6–8 egg chambers. While none of the developing oocytes examined were positive for γ-H2Av staining from control–White depletion, 79% of developing oocytes possessed γ-H2Av signals in female flies lacking Ago3 and Aub proteins in germ cells (Figure 5A and S6A). Similarly, I-element activation in invaded progeny resulted in 72% of oocytes being γ-H2Av positive, versus only 2% in protected control offspring (Figure 5B and S6B). In developing oocytes, chromosomes arrested at the prophase I stage of meiosis were typically packaged within a subnuclear structure known as the karyosome, which appears as a round condensed structure (Mahowald, 1972; Mahowald and Tiefert, 1970). In control ovaries, 95% of developing oocytes had a normal condensed karyosome structure (Figure S6C). However, loss of Ago3&Aub in germ cells made 66% of developing oocytes had defective karyosomes that appeared either stretched or fragmented (Figure S6C). This further suggests that massive retrotransposition events trigger catastrophic genome instability in oocytes. The presence of DNA breaks and defective karyosomes in the nuclei of developing oocytes indicates that these selective retrotransposition events occur within egg chambers undergoing oogenesis.
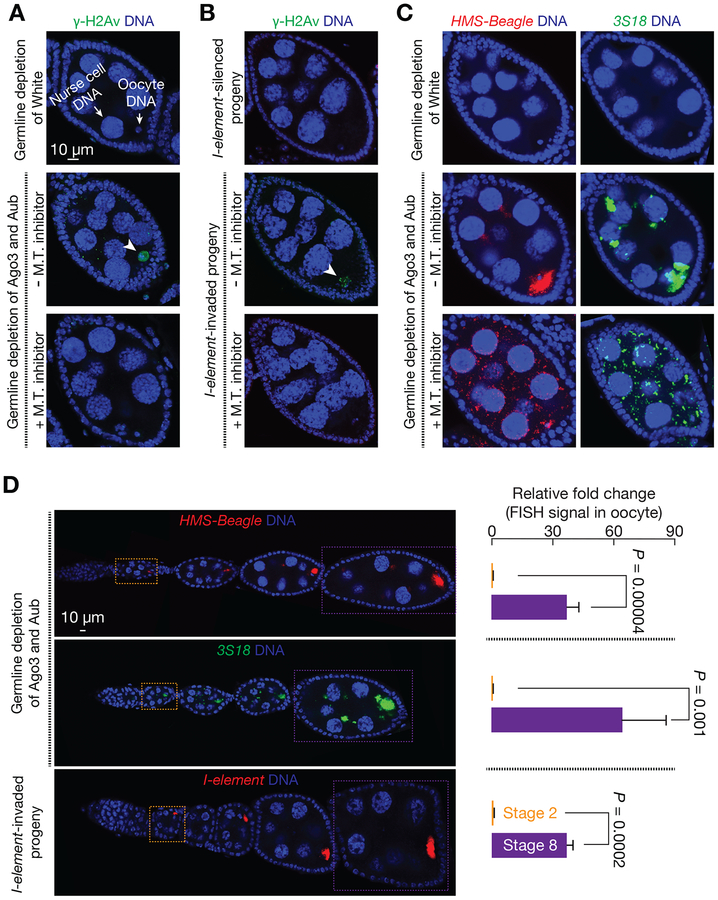
Figure 5. Role of microtubule-mediated transport in selective retrotransposition.
(A and B) γ-H2Av staining to probe the amount of DNA damage in oocyte upon Ago3&Aub depletion (A) or I-element invasion (B). Arrowhead points the oocyte that possesses DNA breaks. Depolymerizing microtubule (M.T.) by Demecolcine-feeding alleviates the DNA damage in oocyte. Data quantification is presented in Figure S6A and S6B.
(C) RNA-FISH to detect HMS-Beagle or 3S18 transcripts upon microtubule depolymerization.
(D) FISH signal quantification to test the potential contribution of early oocyte transcription on the signal detected in the oocytes of stage 8 egg chambers. FISH signal in the oocytes of stage 2 egg chambers defines the maximum amount of mRNAs generated from early stage oocyte. Since there is no oocyte transcription from stage 2 to stage 8. More than 97% of mRNAs in the oocytes of stage 8 egg chambers are transported from nurse cells. Data are represented as mean ± SD. n = 6 ovarioles from 3 animals.
Microtubule-mediated transport delivers transposon materials into oocyte
Because oocytes are transcriptionally inactive (King and Burnett, 1959; Mahowald, 1972; Mahowald and Tiefert, 1970), the factors that support oocyte development are produced in interconnected supporting cells, the nurse cells, and then transported into the oocyte through microtubule (Mahajan-Miklos and Cooley, 1994; Theurkauf et al., 1993). Previous studies have shown that the transcripts from a few transposons, including I-element and 3S18, are enriched in oocytes (Brennecke et al., 2008; Orsi et al., 2010; Seleme et al., 2005; Senti et al., 2015; Shpiz et al., 2007; Tiwari et al., 2017; Van De Bor et al., 2005). We next tested the possibility that retrotransposons selectively target the oocyte genome by “hijacking” the same transportation system. This model predicts that, in the absence of piRNA protection, transposon mRNAs would enrich in oocytes in a microtubule transport-dependent manner. Supporting this model, single-molecule RNA fluorescent in situ hybridization (RNA-FISH) assay reveals strong enrichment of HMS-Beagle transcripts in developing oocytes upon depleting Ago3&Aub in nurse cells (Figure 5C and 5D). Consistent with previous findings (Senti et al., 2015), similar enrichment was detected for 3S18 transcripts (Figure 5C and 5D). By feeding Ago3&Aub depleted flies Demecolcine, which blocks microtubule-dependent transportation (Theurkauf et al., 1993) (as evidenced by the loss of Gurken enrichment in oocyte, Figure S6D), the strong signal of transposon mRNAs from oocytes dramatically diminished (Figure 5C and S2). This indicates that transportation of transposon products depends on the stability of the microtubule network.
Besides importing transcripts from nurse cells, it is possible that oocytes can transcribe transposon mRNAs at very early stage of oogenesis–before chromosome condensing into karyosome, contributing to the signal detected in the later stage egg chambers. To calculate the extent of this potential contribution, we quantified the RNA abundance from the oocytes of stage 2 and stage 8 egg chambers (Figure 5D). Since the oocytes of stage 2 egg chamber already become transcriptionally inert (King and Burnett, 1959; Mahowald, 1972; Mahowald and Tiefert, 1970), the RNA from them reflects the maximum level of contribution from early transcription. Compared to this early stage, the FISH signal in the oocytes of stage 8 egg chambers increased 37-fold for HMS-Beagle, 64-fold for 3S18, and 37-fold for I-element (Figure 5D). Therefore, even if there is no turnover for transposon transcripts in oocyte during oogenesis, the mRNAs from early oocyte transcription would only account for < 3% of the total abundance in the oocytes of stage 8 egg chambers. Our data suggest that the majority of transposon transcripts used for attacking the genome of oocyte are transported from interconnected nurse cells. Consistently, depolymerizing microtubules significantly reduced the presence of DNA breaks and defective karyosomes from oocytes without the protection of piRNAs (Figure 5A, 5B, S6A, S6B, and S6C), further suggesting that retrotransposons preferentially target the oocyte genome by transporting invading materials from nurse cells, via microtubule network.
Neither abundance nor localization of transposon transcripts faithfully reflects mobility
Blocking the piRNA pathway results in dramatic elevation of transcript abundance for the vast majority of transposon families, as measured by extracting total RNA from the whole ovary (Senti et al., 2015). Among the 89 transposon families that potentially have at least 2 full-length copies in the parental genome (Senti et al., 2015), 32 increased their mRNA levels by more than 10-fold upon Ago3&Aub depletion, relative to White depletion; 46 increased by more than 5-fold; and 71 increased by more than 2-fold (Figure 3C and S7A). Based on the dramatic increase of mRNA abundance, it is tempting to conclude that most transposons become active. However, none of them made distinguishable amount of mobilization in ovaries by comparing the genomic sequencing data from Ago3&Aub depleted animals with controls (Figure 3A). Examining transposition in oocyte reveals that only a small group of transposons actively mobilizes even upon piRNA pathway perturbation (Figure 3). Therefore, the relative increase of transcript abundance is not a reliable indicator of mobilization.
Since it is the absolute abundance of transposon mRNAs, rather than the fold-change relative to controls, determines the number of transposases produced. We next tested whether the abundance of transcripts could faithfully reflect the mobile rate of transposons. diver transcribed 1.5-fold more mRNAs than HMS-beagle, but was 11-fold less active at the mobilization level (Figure 3C and S7A). For diver, we did not detect significant increase of transposition upon piRNA pathway perturbation (Figure 3A). In Ago3&Aub depleted ovaries, despite that copia producing similar number of transcripts as Max-element, and 5-fold more than I-element, there was no detectable mobilization in oocytes or ovaries (Figure 3C and S7A). Additionally, we noticed that even having a high amount of mRNAs accumulated in oocyte could not indicate the active mobilization of transposons. Depleting Ago3&Aub led to Burdock produce twice amount of mRNAs than I-element (Figure 3C and S7A), which also enriched in oocytes via microtubule transport (Figure S7B). However, Burdock appears to be nearly inactive on making transposition in oocytes (~1 insertion/genome, Figure 3C and S7A). Based on these findings, we conclude that the abundance of transposon transcripts, often used to define transposon activity, does not faithfully reflect mobilization rate. Our results also suggest the existence of other unknown pathways that suppress transposon mobilization in germ cells.
Retrotransposons rarely mobilize in germline stem cells and nurse cells
Besides selectively targeting oocytes, transposons can also mobilize in the undifferentiated germ cells–primordial germ cells during embryonic and larval stages and germline stem cells in later stages, to achieve heritable mobilization. This is indeed the strategy taken by a DNA transposon, P-element (Teixeira et al., 2017). However, our data indicate that retrotransposons do not mobilize prior to differentiation. Since the differentiated nurse cells and oocytes are derived from the same ovarian stem cells or primordial germ cells, both nurse cells and oocytes would have inherited the same transposition events if they had occurred before differentiation. However, the results from our eGFP reporter showed that only oocytes, neither nurse cells nor germline stem cells, are targeted by I-element for integration (Figure 1). Sequencing the DNA from whole ovaries also suggested that nurse cells have very few, if any, new insertions from I-element and other retrotransposons (Figure 2B, and 3A).
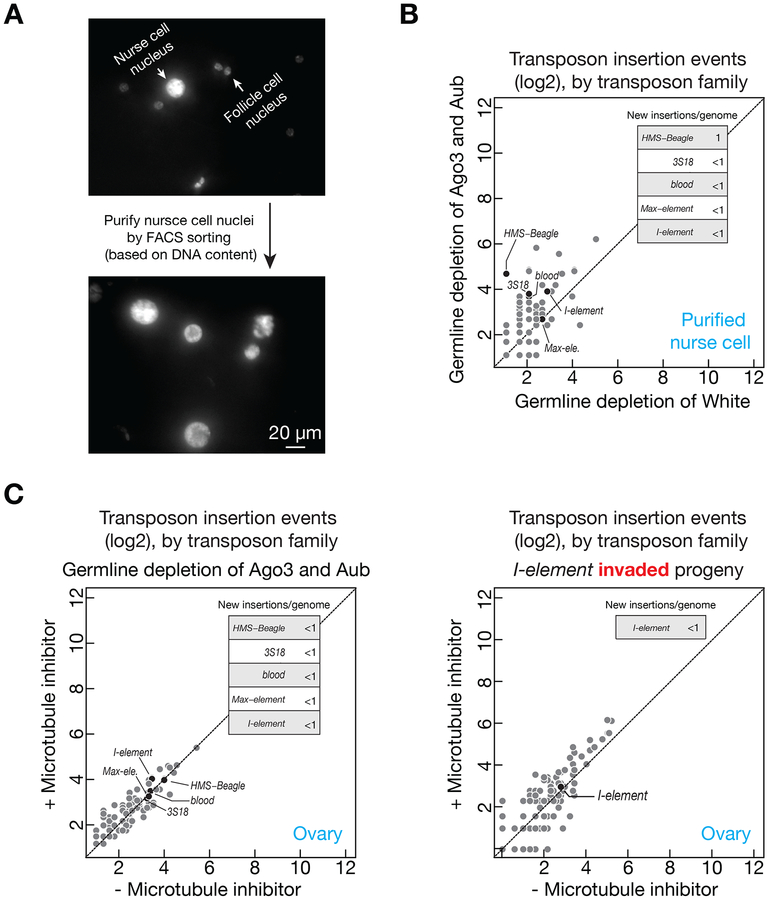
Given that half of the total ovarian DNA is from somatic-lineage follicle cells, to rigorously test the occurrence of transposition in nurse cells, we employed FACS sorting to purify the nuclei of nurse cells and sequenced their genome (Figure 6A). Consistently, for the transposons that made significant amount of integrations in oocytes (Figure 3A and 3B), we only detected ≤ 1 new insertion per genome in nurse cells (Figure 6B, HMS-Beagle is the only one made 1 insertion per genome, all of the rest are < 1). These data reveal that the mobilizations of retrotransposons preferentially occur in oocytes, comparing to nurse cells. Hence, the nurse cell/oocyte integration-dichotomy we observed indicates that retrotransposons unlikely target undifferentiated precursor cells for propagation.
Figure 6. Retrotransposons rarely mobilize in nurse cells.
(A) FACS sorting to purify the nuclei of nurse cells for DNA sequencing. In Drosophila ovaries, the genome of somatic cells (most of them are follicle cells) can only reach 32C. In contrast, nurse cells can achieve 1024C via endo-replication. We have purified the nuclei of cells that have ⩾ 64C DNA.
(B) Upon depleting Ago3&Aub, the number of potential new insertions detected from the DNA of sorted nurse cell. Without the protection from piRNAs, few new insertions were detected in nurse cells.
(C) Either upon depleting Ago3&Aub (left panel) or I-element-invasion (right panel), the number of potential new insertions detected when the transposon mRNAs are restricted within nurse cells (by blocking microtubule-mediated transport). Transposons barely target nurse cell for mobilization.
Being used as factories to manufacture transposon products, it is intriguing that retrotransposons rarely mobilize into nurse cells. This can be explained by the observation that the newly produced transposon transcripts are efficiently transported into oocytes via microtubule (Figure 5C), leaving less transposon products present in nurse cells for mobilization. Additionally, nurse cells may resist to transposition, i.e., transposons might lack the ability to integrate into nurse cell DNA. To investigate, we sequenced ovarian DNA from the Aub&Ago3-depleted or I-element-invaded flies that were fed with microtubule inhibitor, by which the transposon products are restricted within the nurse cells. Upon transposon hyper-activation, although inhibitor feeding retained transposon transcripts within nurse cells (Figure 5C), we did not detect an increase of transposon mobilization events in these cells (Figure 6C). These data indicate that nurse cells are refractory to transposon mobilization.
DISCUSSION
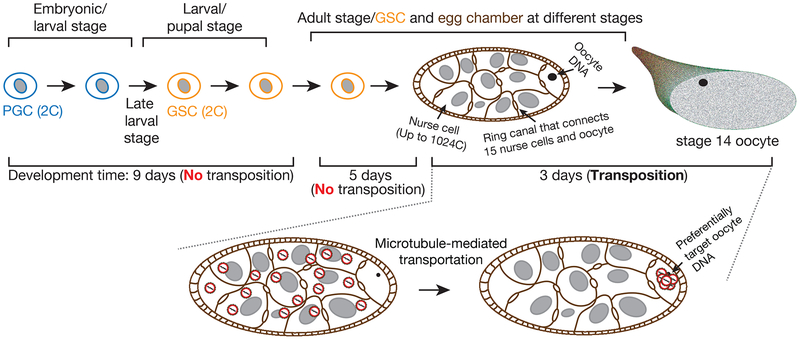
Since somatic transposition is not heritable, transposons must target germ cells to thrive during evolution. Given the unique role of oocyte on passing genetic information, only mobilizations existed in the oocyte, but not other ovarian cells, will transmit to the next generation. To achieve this, transposons can either target undifferentiated stem cells, which later give rise to oocytes, or directly mobilize into developing oocytes, or both. Previous work often uses the transcripts from whole tissue to infer transposon activity (Klattenhoff et al., 2009; Li et al., 2009; Zhang et al., 2011). Although previous studies examined the localization of transposon transcripts during oogenesis by RNA-FISH (Brennecke et al., 2008; Orsi et al., 2010; Seleme et al., 2005; Senti et al., 2015; Shpiz et al., 2007; Tiwari et al., 2017; Van De Bor et al., 2005), there is no reliable information on the activity at the mobilization level, the step that creates mutations and DNA damage. Therefore, little is known on how transposons utilize developmental regulation to achieve the heritable mobilizations. By tracking mobilization with temporal and single-cell resolution, we report that a group of retrotransposons can exploit a short and specific period of fly oogenesis process to efficiently achieve massive propagation by preferentially targeting the oocytes (Figure 7).
Figure 7. A model to depict the transposon mobilization process with spatiotemporal resolution during oogenesis.
Our data indicate that retrotransposons do not mobilize in primordial germ cell (PGC) and germline stem cell (GSC). Instead, within a short and specific time window of fly oogenesis, they take a highly efficient strategy to achieve massive propagation. Likely taking advantage of the polyploid character of nurse cells, these parasitic elements massively manufacture their invading products from up to 15,360 copies of the genome (up to 1024 copies/nurse cell, 15 nurse cells in total within one egg chamber). Instead of mobilizing back into the genome of nurse cells, these invading products are transported into oocyte to selectively target the DNA of oocyte, the only cell that gives rise to the next generation.
Transposon-silencing machinery in germ cells: piRNAs and beyond
With the known function of silencing transposons during gametogenesis, the piRNA pathway has been proposed to lie at the heart of the defending system that protects the hosts from parasitic genomic elements (Aravin et al., 2006; Girard et al., 2006; Grivna et al., 2006; Ishizu et al., 2012; Saito et al., 2006; Siomi et al., 2011; Vagin et al., 2006; Weick and Miska, 2014). Supporting this view, mutating the piRNA pathway factor often results in the drastic increase of the mRNA abundance for most transposon families (Senti et al., 2015). Our data show that, among these families, however, only a small portion can indeed mobilize. By examining the mobility, our results therefore deliver a surprising message: the vast majority of transposon families still could not transpose even upon the piRNA pathway perturbation. Perhaps they have acquired mutations that terminate their mobility during evolution. Although might be true for some families, this unlikely explains all of them. Accumulating evidence suggests that most transposons in fly genome are still, although at a low frequency, capable of mobilizing, contributing to 79% of large genetic variation (Chakraborty et al., 2018; Kaminker et al., 2002). Examples include: Doc recently transposed into the white locus generated the first documented white-eye fly (Driver et al., 1989; Morgan, 1910); Accord mobilization contributes to the tandem duplication of the genes associated with nicotine resistance (Chakraborty et al., 2018); mdg3 inserted into the 3’UTR of mRpL48 and fritz (Chakraborty et al., 2018); copia mobilized into tonally (Chakraborty et al., 2018). Although these families are all targeted by the piRNA machinery (Senti et al., 2015), we barely detect any mobilization for them in all samples we examined, even upon blocking the piRNA pathway. Altogether, these observations suggest that in addition to the piRNA pathway, the germ cells employ other mechanisms to suppress transposons from massive propagation.
Transposon propagation strategy
For transposons that massively propagate upon loss of piRNA protection, we did not detect mobilization in undifferentiated germ cells: primordial germ cells and germline stem cells. This may reflect that these undifferentiated germ cells still could achieve transposon silencing even without the protection from piRNAs. Alternatively, retrotransposons likely evolved to avoid these cells for targeting since their diploid character limits them to only two copies of the genome for the production of transposon products. Instead, after germline stem cell differentiation, retrotransposons are unleashed from the genomes of supporting nurse cells, which are highly polyploid (up to 1024C) (Dej and Spradling, 1999; Painter and Reindorp, 1939). However, since these nurse cells will eventually die at the end of oogenesis (Buszczak and Cooley, 2000), it would be futile for transposons to target the genome of nurse cells. Our study suggests that retrotransposons mainly utilize nurse cells as factories to massively produce the invading products, but rarely transpose into their genome (Figure 7). Rather, these selfish elements hijack the oogenesis process and selectively mobilize into the genome of the interconnected neighbor cells, oocytes–the founders of the next generation (Figure 7). Within one egg chamber, since the ratio of nurse cells to oocyte is 15:1, retrotransposons could amplify from up to 15,360 (1024×15) copies of the genome before mobilizing into the oocyte DNA. Given this intensive pressure that can lead to catastrophic genome instability and embryonic lethality, our finding explains why Drosophila has to evolve a specialized small RNA-based “immune” system, the piRNA pathway, to largely, but nearly impossible to completely, suppress transposable elements for species survival.
Recent findings suggest that oogenesis in mammals shares significant amount of similarities as Drosophila (Lei and Spradling, 2016). Mammalian primordial germ cells give rise to germline cysts or syncytia, which include nurselike neighbor cells and developing oocytes (Grive and Freiman, 2015). Moreover, the key steps that support Drosophila oogenesis–membrane breakdown within the cyst, apoptosis process, and intercellular transport–also appear to play an important role in mammalian oocyte development (Lei and Spradling, 2016). Therefore, retrotransposons may use a similar scheme to accomplish cell-type-specific targeting. Supporting this idea, the LINE1 transposons from mouse genome indeed show differential accumulation of the Orf1p proteins among otherwise genetically identical cells within a cyst (Malki et al., 2014). Additionally, accumulating evidence indicates that retrotransposon activation contributes to mammalian embryogenesis, neurogenesis, and tumorigenesis (Burns, 2017; Erwin et al., 2016; Grow et al., 2015; Kazazian and Moran, 2017). Perhaps these endogenous retroviruses drive development or promote disease also by taking their preferential rides.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Zhao Zhang (zzhang@carnegiescience.edu)
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Fly strains, housing and husbandry conditions
The following fly lines were used: w1118 (Brennecke et al., Science, 2008), wk (Brennecke et al., Science, 2008), MTD-Gal4: P(otu-Gal4::VP16.R)1 w[*]; P(Gal4-nos.NGT)40; P(Gal4::VP16-nos.UTR)CG6325[MVD1], white-sh (attP2) (Senti et al., Genes & Dev., 2015), aub-sh (attP40) and ago3-sh (attP2) (Senti et al., Genes & Dev., 2015), X-Y attached male: c(1;Y)1,yw/O, w1118-I-element-eGFP-reporter (this study).
All flies were maintained at 25°C and grown on standard agar-corn medium. For the crosses to study I-element activation, all parental flies were less than 8-day-old. The spermless males (XO) were produced by crossing X-Y attached males with virgin females. Flies with the age of 3–7 days were used for FISH and immunostaining. The F1 generation eggs, which serve as controls to define transposon content from parental strains, were collected within 3 hours after being laid. Overnight oocyte collection was performed for the F1 females mated with spermless males. To get the flies without the protection of piRNA pathway, we chose RNAi-based protein depletion over genetic mutation for the following two reasons: 1), the mutants are generated from different genetic background, and may differ dramatically with the transposon contents; 2) likely caused by the accumulation of secondary mutations, the mutant flies lay very few eggs, therefore it is impossible to collect adequate number of oocytes from them.
METHOD DETAILS
eGFP reporter
The DNA fragment that contains eGFP-reporter and I-element 3’UTR was synthesized. The I-element (without 3’UTR) was amplified from PI407 vector (kind gift from Dr. Silke Jensen), and cloned into the EcoRI and NotI sites of pCaSpeR3 vector to generate pCaS3-I-X3UTR. Stbl2 (ThermoFisher, Cat#: 10268019) competent cells were used for this cloning and the plates have to be left at room temperature for correct colonies to grow. The eGFP-reporter-I-element-3’UTR fragment was cloned into the SphI and AscI sites (engineered through PCR primer) of pCaS3-I-X3UTR to get the I-element transposition reporter construct and transformed into Stbl4 competent cells (ThermoFisher, Cat#: 11635018).
To detect the eGFP signal, 2–4 day old female flies were fed with yeast paste for overnight. The flies were incubated in 37°C water bath for 30 minutes, and recovered at 25°C for 3–5 hours. The ovaries were dissected to check eGFP expression.
Genomic library preparation
Genomic DNA from different tissues was extracted by using Quick-gDNA MicroPrep kit (ZYMO RESEARCH, Cat #D3021). After extraction, 12.5 ng of genomic DNA was used for genomic library preparation by using Illumina Nextera DNA Library Preparation kit (FC121–1030 and FC121–1011). Briefly, 12.5 ng (in 10 μl volume) of genomic DNA was mixed with 12.5 μl of TD Buffer and 2.5 μl of TDE1. On a PCR machine, the mixture was incubated at 55 °C for 10 minutes. Then 25 μl water and 250 μl DNA binding buffer (from ZYMO DNA Clean and Concentrator-5, Cat #: D4004) was added to purify the DNA with the Concentrator-5 kit. The DNA was eluted with 12.5 μl of RSB. One μl of the sample was saved for Bioanalyzer analysis. For the rest sample, 2.5 μl of Index 1(i7), 2.5 μl of Index 2 (i5), 7.5 μl NPM, and 2.5 μl PPC was added and mixed well. On a PCR machine, the mixture was incubated at 72°C for 3 min, 98°C for 30 sec, and 7 cycles of the following steps: 98°C for 10 sec, 63°C for 30 sec, 72°C for 3 min. Finally, 25 μl water was added to the PCR reaction, then 30 μl of Ampure XP beads was used to purify the DNA. The library was eluted with 20 μl EB. The library was sequenced by Illumina NextSeq machine with the run of Paired-End 75 nt.
Note, we noticed that the traditional DNA library preparation method, which involves DNA fragmentation, End-repair, Adapter-ligation, appears to produce high false-positive rates for detecting potential new transposon insertions. We suspect that this is caused by the chimeric DNA fragments generated by the ligation step. Therefore, we used the Tn5 tagmentation approach (Nextera kit), which avoids the ligation step, to generate all of the genomic libraries for this study.
High throughput Sequencing Data analysis
The genomic libraries were first mapped to dm6 genome to estimate the library mappability by BWA v0.7.15. Because of the DNA extracted from the oocytes that purified by crossing with spermless males contains large fraction of mitochondria genome (and wolbachia genome for the samples related with I-element study), we next randomly selected the corresponding number of reads from each library to achieve the same genome coverage for comparison. The selected reads were re-mapped to dm6 genome by BWA. The reads pair information for each sequenced fragment was obtained by bwa sampe. The sorted bam output from BWA mapping was funneled to detect new transposon insertions by TEMP (Zhuang et al., 2014). The potential new insertions that also appear in the F1 egg samples (representing the parental genome) were discarded. Only transposon families that have more than two copies per genome were used for plotting with 5 pseudocounts added. Whether the transposons are predicted to be capable of making autonomous mobilization is referred from the previous publication (Kaminker et al., 2002). The transposon families that were not analyzed from this paper are manually checked by searching the full-length copy from Drosophila melanogaster genome (dm6) by using UCSC blat (https://genome.ucsc.edu/cgi-bin/hgBlat?command=start).
We noticed that w1118 flies have heavy wolbachia contamination, which maternally passes into I-element-silenced control oocytes. This could potentially be problematic when the spermless males were used to purify oocyte DNA. Since the oocytes could not develop (lacking of sperm DNA), it leads to a large fraction of sequencing reads were from wolbachia genome. To rigorously measure the potential transpositions in control oocytes, we have additionally sequenced the embryos laid by mating the I-element-silenced control females with wk males, which allows embryos to develop for 2–3 hours and dilute the relative content of wolbachia DNA. Given the DNA from wk male contributes to half of the genome from these egg, we have used twice number of reads for the analysis.
Transposon transcript abundance from previously published strand-specific RNA-Seq data (Senti et al., 2015) was calculated by first mapping reads using TopHat v2.1.1 to the full length consensus sequences for transposable elements described in (Senti et al., 2015). Raw counts were calculated for each of the 122 transposons, with multi-mapping reads assigned proportionally. Counts were normalized using size factors calculated by DESeq analysis of the same reads mapped to the BDGP6 gene set annotated in Ensembl release 85. Finally, normalized counts were converted to normalized ppm by dividing with the average number of BDGP6 mapped reads across the datasets. The flies used for RNA-Seq and DNA-Seq have the same genetic background.
For hotspots analysis, Drosophila genome (without Y chromosome) was split into 13391 bins by taking 10 kb as the window size by using “bedtools makewindows”. The theoretical number of insertions per bin was calculated by assuming there is not landing preference for the transposons analyzed. Then the number of unique detected insertions per bin for each transposon was calculated by using “bedtools intersect”. We define the bins that have 10 times greater insertions than the theoretical number as potential hotspots. The hotspots that adjacent to each other were merged by “bedtools merge” (Table S1). The circos plot to display all hotspots was generated by the R package–circlcize: circus.trackHist (Gu et al., 2014).
RNA-FISH
Stellaris RNA FISH probe sets for HMS-Beagle, 3S18, I-element were designed and purchased from LGC Bioresearch Techlologies, and the probe sequences were listed in Table S2. Briefly, 3 pairs of ovaries were dissected in cold PBS and fixed for 20 minutes in 4% formaldehyde. Ovaries were washed once with PBST and twice with PBS, and then immersed in 70% (v/v) ethanol for 8 hours at 4°C. After that, ovaries were washed once with Wash Buffer A (LGC Biosearch Tech, Cat# SMF-WA1–60) at room temperature for 5 minutes, then incubated with 50 μl Hybridization Buffer (LGC Biosearch Tech, Cat# SMF-HB1–10) containing probe set (125 nM) for hybridization at 37 °C for overnight. Next day, ovaries were washed twice with Wash Buffer A for 30 minutes at 37 °C and once with Wash Buffer B (LGC Biosearch Tech, Cat# SMF-WB1–20) for 5 minutes at room temperature. Samples were mounted with 20 μl Vectashield Mounting Medium (VECTOR LABORATORIES INC MS, 101098–042). For signal quantification, ~30 Z-stack images of individual sample were taken by the Leica TCS SP5 confocal microscope. The florescence signal from stage 2 and stage 8 egg chambers was determined by ImageJ. For each oocyte, the FISH signal was calculated by subtracting the brightness of the background (blank region with the same area) from the total brightness determined by ImageJ.
Immunostaining
Briefly, 3 pairs of ovaries were dissected in cold PBS and fixed for 20 minutes in 4% formaldehyde. Ovaries were washed 3 times with PBST, then incubated with blocking solution containing 2% BSA and 2% NGS at room temperature for 1 hour. Primary antibodies were diluted in blocking solution and incubated at 4 °C for overnight. Ovarioles were washed 3 times with PBST for 10 minutes, and then incubated with secondary antibody at room temperature for 2 hours. Ovary samples were washed 3 times with PBST for 10 minutes and stained with DAPI for 8 min, followed by washing 2 times with PBST for 10min, finally mounted with 20 μl Vectashield Mounting Medium. Egg chambers from stage 6–8 were imaged and quantified. Primary antibodies used in this study were: anti-Aub (1:50), anti-Ago3 (1:50), anti-γH2Av (1:400), anti-Vasa (1:5,000), anti-Gurken (1:100). Secondary antibodies were used at 1:400 and chromatin was visualized by DAPI.
Microtubule assembly inhibitor treatment
Briefly, 2–4 day female files raised at 25°C on a standard corn-meal agar diet were starved for 12 hours, then these flies were fed with yeast paste supplemented with 50μg/ml Demecolcine (Sigma, D7385) for 24 hours. Three pairs of ovaries were dissected for DAPI staining, immunostaining or RNA FISH. Egg chambers from stage 6–8 were imaged and quantified. Note: Demecolcine treatment made ~25% egg chambers have 16 nurse cells without oocyte, as expected (Theurkauf et al., 1993).
Nurse cell purification and genomic DNA extraction
Briefly, 300 female flies (0–2 day) were collected and fed with wet yeast paste for 2 days. Ovaries were dissected in cold PBS in batches of ~60 flies and then treated with collagenase (5 mg/ml) for 10 minutes. During collagenase treatment, the samples were pipetted up and down every 3 minutes to break up ovarioles. Ovary samples were washed once with PBS and then fixed with 4% PFA for 10 minutes at room temperature, then neutralized with 2.5M glycine for 5 minutes. Next, filtering stage 14 egg chambers away and then washing samples twice with PBS (Cells can be frozen in liquid nitrogen and placed at −80 degrees until ready for FACS sorting). Before Sorting, cell clusters and un-disassociated egg chambers were treated with 0.125% trypsin for 10 minutes at room temperature, then washed twice with PBS. All cells were Re-suspended in 0.5 ml of NIB (15 mM Tris-HCl pH 7.4, 60 mM KCl, 15 mM NaCl, 250 mM sucrose, 1 mM EDTA, 0.1 mM EGTA, 0.15mM spermine, 0.5mM spermidine, 1.5% NP40) plus DAPI and transferred to 2 ml dounce homogenizer. Samples were dounced 45 times with pestle B. Then, cells were placed on top of sucrose step (0.5ml 2M sucrose/0.5ml 1M sucrose) and spun 20 minutes with 20000 g at 4 degrees. After spin, removing all liquid and re-suspending cells in 1ml of NIB. Nurse cells that have more than 64C DNA were sorted out by FACS. Purified nurse cells were spun 5 minutes at 1000g. After removing all liquid, samples were treated by adding 80 μl of 2× proteinase K buffer and 3 μl of 5M NaCl, then were incubated at 65 °C for overnight. Next day, 1 μl of 32 mg/ml RNase A was added into each tube and incubate all samples at 37 °C for 30 minutes. Next, cells were digested with 20 mg/ml Proteinase K at 55 °C for 2 hours. DNA was then extracted by Phenol/Chloroform and precipitated by ethanol. A detailed protocol is available upon request.
QUANTIFICATION AND STATISTICAL ANALYSIS
Student’s T test was carried out using either Microsoft Excel or R to calculate significance. P values are indicated on plots. Results are expressed as mean ± standard error (SD). The number of animals examined are indicated in figure legends.
DATA AND SOFTWARE AVAILABILITY
All sequencing data generated from this study are available at NCBI: SRP144483.
Supplementary Material
Table S1. Hotspots for HMS-Beagle, blood, 3S18, and I-element. Related to Figure 4
Table S3. Summary of genomic libraries. Related to STAR methods
Figure S1. Monitoring transposition with an eGFP reporter indicates I-element preferentially inserts into oocyte genome, Related to Figure 1
(A) Schematic design of eGFP reporter to track I-element mobilization.
(B) Fly cross scheme to collect samples for measuring the potential transposition events from invaded and control flies.
(C) Probing transposition events by PCR. Transpositions in oocytes give rise to the PCR product without the intron. Primers are depicted in panel A. Samples were boxed with different color to match the corresponding materials depicted in panel B.
Figure S2. Upon activation, I-element mRNAs are enriched in oocytes in a microtubule-dependent manner, Related to Figure 1 and Figure 5
RNA-FISH to detect the I-element transcripts in invaded ovaries. The enrichment is abolished when the microtubule (M.T.)-mediated transport is blocked (Demecolcine treatment). Data are represented as mean ± SD. The egg chambers examined for each condition are from 9 animals.
Figure S3. Globally measure transposon mobilization in I-element-invaded progeny, Related to Figure 2
(A) Fly cross scheme.
(B) Scatter plot to display the detected potential new insertions.
Figure S4. Fly cross scheme to globally measure transposon mobilization upon Ago3&Aub depletion in ovaries, Related to Figure 3
(A) Fly cross scheme.
(B) Immuno-staining pictures to demonstrate the RNAi efficiency for Ago3 and Aub depletion.
Figure S5. Loss of Ago3&Aub leads to transposon insertions accumulated in oocytes, Related to Figure 3
(A) Genome sequencing to globally probe new transposition events from carcass samples, which serve as negative control.
(B) The new transposition events from oocytes were plotted to demonstrate both fold change and P-value for each transposon family. GLD = Germline Depletion.
(C) The number of detected new insertions shows linear correlation with the fold of genome coverage.
Figure S6. Blocking microtubule-mediated transport alleviates DNA damage in oocyte upon retrotransposon activation, Related to Figure 5
(A and B) Quantification of oocytes that do not have DNA damage signal upon piRNA pathway perturbation (A) or I-element invasion (B). GLD = Germline Depletion. Note, since we still count the oocytes that have weak γ-H2Av signals as DNA damage positive from Demecolcine-treated animals, we most likely have underestimated the rescue phenotype upon blocking microtubule transport. Data are represented as mean ± SD. The oocytes examined for each condition are from 9 animals.
(C) DAPI staining to measure karyosome morphology. Transposon mobilization in oocytes leads to karyosome packing defects that can be rescued by depolymerizing microtubule. In normal oocyte, the DNA is packed into a round condensed structure, named karyosome. Depleting Ago3 and Aub in germ cells results in karyosome packing defects (either stretched or fragmented). Because blocking microtubule-mediated transport made only 54% of karyosomes from control animals (White-depleted) are normal, depolymerizing microtubule thus appears to rescue the defects in Ago3&Aub depleted ovaries to control level (51%). GLD = Germline Depletion. Data are represented as mean ± SD. The oocytes examined for each condition are from 9 animals.
(D) Gurken staining to validate the effect of Demecolcine on microtubule-mediated transport.
Figure S7. Neither abundance nor localization of transposon mRNAs reflects mobility, Related to Figure 3 and Figure 7
(A) Scatter plots to display the number of new insertions detected in oocytes and the abundance of transposon mRNAs in ovaries. GLD = Germline Depletion.
(B) RNA-FISH to detect the localization of Burdock mRNA. Abundant Burdock mRNAs enrich in oocytes in the microtubule-dependent manner, but Burdock rarely mobilizes.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-Aub | Senti et al., 2015 | N/A |
| Mouse monoclonal anti-Ago3 | Senti et al., 2015 | N/A |
| Mouse monoclonal anti-γH2Av | DSHB | CAT#: UNC93–5.2.1 |
| Rabbit polyclonal anti-Vasa | Lasko et al., 1990 | N/A |
| Mouse anti-Gurken | DSHB | CAT#: 1D12-s |
| Alexa 488 Donkey anti-Mouse | Molecular Probes | CAT#: A21202 |
| Alexa 594 Goat anti-Rabbit | Molecular Probes | CAT#: A11037 |
| Experimental Models: Organisms/Strains | ||
| D.melanogaste: w1118 | Brennecke et al., 2008 | N/A |
| D.melanogaster: wk | Brennecke et al., 2008 | N/A |
| D.melanogaster: P(otu-Gal4::VP16.R)1 w[*]; P(Gal4-nos.NGT)40; P(Gal4::VP16-nos.UTR)CG6325[MVD1] | Bloomington Drosophila Stock Center | CAT#: 31777 |
| D.melanogaster: white-sh (attP2) | Senti et al., 2015 | N/A |
| D.melanogaster: aub-sh (attP40) and ago3-sh (attP2) | Senti et al., 2015 | N/A |
| D.melanogaster: X-Y attached male: c(1;Y)1,yw/O | Robert Levis (Spradling lab) | N/A |
| D.melanogaster: w1118-I-element-eGFP-reporter | This paper | N/A |
| Competent cells | ||
| Stbl2 | ThermoFisher | CAT#: 10268019 |
| Stbl4 | ThermoFisher | CAT#: 11635018 |
| Critical Commercial Assay | ||
| Nextera DNA Library Prep | Illumina | CAT#: FC-121–1030 and FC121–1011 |
| Quick-gDNA MicroPrep | Zymo Research | CAT#: D3021 |
| DNA Clean and Concentrator-5 | Zymo Research | CAT#: D4004 |
| FISH Wash Buffer A | LGC Biosearch Tech | CAT#: SMF-WA1–60 |
| FISH Hybridization Buffer | LGC Biosearch Tech | CAT#: SMF- HB1–10 |
| FISH Wash Buffer B | LGC Biosearch Tech | CAT#: SMF- WB1–20 |
| Vectashield Mounting Medium | VECTOR LABORATORIES INC MS | CAT#: 101098–042 |
| Demecolcine | Sigma | CAT#: D7385 |
| Deposited Data | ||
| Illumina Sequencing raw data (FASTQ) | This paper | https://www.ncbi.nlm.nih.qov/bioproiect/PRJNA454868 |
| RNA sequencing data | Senti et al., 2015 | GSE71775 |
| Oligonucleotides | ||
| Forward primer to amplify I-element without 3′UTR: CCG GAA TTC TTA GTA AAT TAT TAA TAT GCA AAT CAT TAC CAC TTC AAC CTC CG AAG | This paper | N/A |
| Reverse primer to amplify I-element without 3′UTR: TTT TCC TTT TGC GGC CGC ACA TGC ATG CAT GTC GAC GCG TCG TTG GCG CGC CAC AAA TGC CTG TTT TGT TTT TAG A | This paper | N/A |
| Forward primer to amplify the eGFP-reporter-I-3′UT: CGT TGG CGC GCC GGT ACC GAA T | This paper | N/A |
| Reverse Primer to amplify the eGFP-reporter-I-3′UTR: CGA TAC ATG CAT GCA AAA CGT CTT TGC CAC GAT T | This paper | N/A |
| Software and Algorithms | ||
| piPipes | Han et al., 2015 | N/A |
| TEMP | Zhuang et al., 2014 | N/A |
| circlcize | Gu et al., 2014 | N/A |
| Other | ||
ACKNOWLEDGEMENTS
We thank Julius Brennecke for providing antibodies and fly alleles, Silke Jensen for sharing I-element plasmid, Robert Levis for X-Y attached fly allele. We thank members from ZZ lab, Spradling lab, and Bortvin lab for critical suggestions, Madeline Cassani for assistance on cloning. We also thank Allan Spradling and Marnie Halpern for giving valuable suggestions on the manuscript. This work was supported by the grant from the National Institutes of Health to Z.Z. (DP5 OD021355) and Carnegie Endowment. All authors, as junior scientists and young parents, sincerely thank our family for their immeasurable love and sacrifice to support us during the story development/publishing process.
Footnotes
DECLARATION OF INTERESTS
The authors declare that they have no conflict of interest.
REFERENCES
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. (2006). A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442, 203–207. [DOI] [PubMed] [Google Scholar]
- Boeke JD, Garfinkel DJ, Styles CA, and Fink GR (1985). Ty elements transpose through an RNA intermediate. Cell 40, 491–500. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, and Hannon GJ (2007). Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128, 1089–1103. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A, and Hannon GJ (2008). An epigenetic role for maternally inherited piRNAs in transposon silencing. Science 322, 1387–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucheton A, Paro R, Sang HM, Pelisson A, and Finnegan DJ (1984). The molecular basis of I-R hybrid dysgenesis in Drosophila melanogaster: identification, cloning, and properties of the I factor. Cell 38, 153–163. [DOI] [PubMed] [Google Scholar]
- Burns KH (2017). Transposable elements in cancer. Nature reviews Cancer 17, 415–424. [DOI] [PubMed] [Google Scholar]
- Buszczak M, and Cooley L (2000). Eggs to die for: cell death during Drosophila oogenesis. Cell Death Differ 7, 1071–1074. [DOI] [PubMed] [Google Scholar]
- Chakraborty M, VanKuren NW, Zhao R, Zhang X, Kalsow S, and Emerson JJ (2018). Hidden genetic variation shapes the structure of functional elements in Drosophila. Nature genetics 50, 20–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanov P, Devroe E, Silver PA, and Engelman A (2004). Identification of an evolutionarily conserved domain in human lens epithelium-derived growth factor/transcriptional co-activator p75 (LEDGF/p75) that binds HIV-1 integrase. The Journal of biological chemistry 279, 48883–48892. [DOI] [PubMed] [Google Scholar]
- Chuong EB, Elde NC, and Feschotte C (2017). Regulatory activities of transposable elements: from conflicts to benefits. Nature reviews Genetics 18, 71–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dej KJ, and Spradling AC (1999). The endocycle controls nurse cell polytene chromosome structure during Drosophila oogenesis. Development 126, 293–303. [DOI] [PubMed] [Google Scholar]
- Dombroski BA, Mathias SL, Nanthakumar E, Scott AF, and Kazazian HH Jr. (1991). Isolation of an active human transposable element. Science 254, 1805–1808. [DOI] [PubMed] [Google Scholar]
- Driver A, Lacey SF, Cullingford TE, Mitchelson A, and O’Hare K (1989). Structural analysis of Doc transposable elements associated with mutations at the white and suppressor of forked loci of Drosophila melanogaster. Mol Gen Genet 220, 49–52. [DOI] [PubMed] [Google Scholar]
- Erwin JA, Paquola AC, Singer T, Gallina I, Novotny M, Quayle C, Bedrosian TA, Alves FI, Butcher CR, Herdy JR, et al. (2016). L1-associated genomic regions are deleted in somatic cells of the healthy human brain. Nature neuroscience. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Moran JV, Kazazian HH Jr., and Boeke JD (1996). Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell 87, 905–916. [DOI] [PubMed] [Google Scholar]
- Girard A, Sachidanandam R, Hannon GJ, and Carmell MA (2006). A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 442, 199–202. [DOI] [PubMed] [Google Scholar]
- Grive KJ, and Freiman RN (2015). The developmental origins of the mammalian ovarian reserve. Development 142, 2554–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivna ST, Beyret E, Wang Z, and Lin H (2006). A novel class of small RNAs in mouse spermatogenic cells. Genes & development 20, 1709–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grow EJ, Flynn RA, Chavez SL, Bayless NL, Wossidlo M, Wesche DJ, Martin L, Ware CB, Blish CA, Chang HY, et al. (2015). Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells. Nature 522, 221–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Gu L, Eils R, Schlesner M, and Brors B (2014). circlize Implements and enhances circular visualization in R. Bioinformatics 30, 2811–2812. [DOI] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, and Siomi MC (2007). A slicer-mediated mechanism for repeat-associated siRNA 5’ end formation in Drosophila. Science 315, 1587–1590. [DOI] [PubMed] [Google Scholar]
- Han BW, Wang W, Li C, Weng Z, and Zamore PD (2015). Noncoding RNA. piRNA-guided transposon cleavage initiates Zucchini-dependent, phased piRNA production. Science 348, 817–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizu H, Siomi H, and Siomi MC (2012). Biology of PIWI-interacting RNAs: new insights into biogenesis and function inside and outside of germlines. Genes & development 26, 2361–2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen S, and Heidmann T (1991). An indicator gene for detection of germline retrotransposition in transgenic Drosophila demonstrates RNA-mediated transposition of the LINE I element. The EMBO journal 10, 1927–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminker JS, Bergman CM, Kronmiller B, Carlson J, Svirskas R, Patel S, Frise E, Wheeler DA, Lewis SE, Rubin GM, et al. (2002). The transposable elements of the Drosophila melanogaster euchromatin: a genomics perspective. Genome biology 3, RESEARCH0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian HH Jr., and Moran JV (2017). Mobile DNA in Health and Disease. The New England journal of medicine 377, 361–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RC, and Burnett RG (1959). Autoradiographic study of uptake of tritiated glycine, thymidine, and uridine by fruit fly ovaries. Science 129, 1674–1675. [DOI] [PubMed] [Google Scholar]
- Klattenhoff C, Bratu DP, McGinnis-Schultz N, Koppetsch BS, Cook HA, and Theurkauf WE (2007). Drosophila rasiRNA pathway mutations disrupt embryonicaxis specification through activation of an ATR/Chk2 DNA damage response. Dev Cell 12, 45–55. [DOI] [PubMed] [Google Scholar]
- Klattenhoff C, Xi H, Li C, Lee S, Xu J, Khurana JS, Zhang F, Schultz N, Koppetsch BS, Nowosielska A, et al. (2009). The Drosophila HP1 homolog Rhino is required for transposon silencing and piRNA production by dual-strand clusters. Cell 138, 1137–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei L, and Spradling AC (2016). Mouse oocytes differentiate through organelle enrichment from sister cyst germ cells. Science 352, 95–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Vagin VV, Lee S, Xu J, Ma S, Xi H, Seitz H, Horwich MD, Syrzycka M, Honda BM, et al. (2009). Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell 137, 509–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan-Miklos S, and Cooley L (1994). Intercellular cytoplasm transport during Drosophila oogenesis. Developmental biology 165, 336–351. [DOI] [PubMed] [Google Scholar]
- Mahowald AP (1972). Ultrastructural observations on oogenesis in Drosophila. J Morphol 137, 29–48. [DOI] [PubMed] [Google Scholar]
- Mahowald AP, and Tiefert M (1970). Fine structural changes in theDrosophila oocyte nucleus during a short period of RNA synthesis : An autoradiographic and ultrastructural study of RNA synthesis in the oocyte nucleus of Drosophila. Wilhelm Roux Arch Entwickl Mech Org 165, 8–25. [DOI] [PubMed] [Google Scholar]
- Malki S, van der Heijden GW, O’Donnell KA, Martin SL, and Bortvin A (2014). A role for retrotransposon LINE-1 in fetal oocyte attrition in mice. Dev Cell 29, 521–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki Y, Nishisho I, Horii A, Miyoshi Y, Utsunomiya J, Kinzler KW, Vogelstein B, and Nakamura Y (1992). Disruption of the APC gene by a retrotransposal insertion of L1 sequence in a colon cancer. Cancer research 52, 643–645. [PubMed] [Google Scholar]
- Mohn F, Handler D, and Brennecke J (2015). Noncoding RNA. piRNA-guided slicing specifies transcripts for Zucchini-dependent, phased piRNA biogenesis. Science 348, 812–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, and Kazazian HH Jr. (1996). High frequency retrotransposition in cultured mammalian cells. Cell 87, 917–927. [DOI] [PubMed] [Google Scholar]
- Morgan TH (1910). Sex Limited Inheritance in Drosophila. Science 32, 120–122. [DOI] [PubMed] [Google Scholar]
- Orsi GA, Joyce EF, Couble P, McKim KS, and Loppin B (2010). Drosophila I-R hybrid dysgenesis is associated with catastrophic meiosis and abnormal zygote formation. Journal of cell science 123, 3515–3524. [DOI] [PubMed] [Google Scholar]
- Painter TS, and Reindorp EC (1939). Endomitosis in the nurse cells of the ovary of drosophila melanogaster. Chromosoma 1, 276–283. [Google Scholar]
- Picard G (1976). Non-mendelian female sterility in Drosophila melanogaster: hereditary transmission of I factor. Genetics 83, 107–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Nishida KM, Mori T, Kawamura Y, Miyoshi K, Nagami T, Siomi H, and Siomi MC (2006). Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes & development 20, 2214–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seleme MC, Disson O, Robin S, Brun C, Teninges D, and Bucheton A (2005). In vivo RNA localization of I factor, a non-LTR retrotransposon, requires a cis-acting signal in ORF2 and ORF1 protein. Nucleic acids research 33, 776–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senti KA, Jurczak D, Sachidanandam R, and Brennecke J (2015). piRNA-guided slicing of transposon transcripts enforces their transcriptional silencing via specifying the nuclear piRNA repertoire. Genes & development 29, 1747–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpiz S, Kwon D, Uneva A, Kim M, Klenov M, Rozovsky Y, Georgiev P, Savitsky M, and Kalmykova A (2007). Characterization of Drosophila telomeric retroelement TAHRE: transcription, transpositions, and RNAi-based regulation of expression. Mol Biol Evol 24, 2535–2545. [DOI] [PubMed] [Google Scholar]
- Siomi MC, Sato K, Pezic D, and Aravin AA (2011). PIWI-interacting small RNAs: the vanguard of genome defence. Nature reviews Molecular cell biology 12, 246–258. [DOI] [PubMed] [Google Scholar]
- Spradling AC (1993). Germline cysts: communes that work. Cell 72, 649–651. [DOI] [PubMed] [Google Scholar]
- Teixeira FK, Okuniewska M, Malone CD, Coux RX, Rio DC, and Lehmann R (2017). piRNA-mediated regulation of transposon alternative splicing in the soma and germ line. Nature 552, 268–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theurkauf WE, Alberts BM, Jan YN, and Jongens TA (1993). A central role for microtubules in the differentiation of Drosophila oocytes. Development 118, 1169–1180. [DOI] [PubMed] [Google Scholar]
- Tiwari B, Kurtz P, Jones AE, Wylie A, Amatruda JF, Boggupalli DP, Gonsalvez GB, and Abrams JM (2017). Retrotransposons Mimic Germ Plasm Determinants to Promote Transgenerational Inheritance. Current biology : CB 27, 3010–3016 e3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, and Zamore PD (2006). A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313, 320–324. [DOI] [PubMed] [Google Scholar]
- Van De Bor V, Hartswood E, Jones C, Finnegan D, and Davis I (2005). gurken and the I factor retrotransposon RNAs share common localization signals and machinery. Dev Cell 9, 51–62. [DOI] [PubMed] [Google Scholar]
- Wang W, Han BW, Tipping C, Ge DT, Zhang Z, Weng Z, and Zamore PD (2015). Slicing and Binding by Ago3 or Aub Trigger Piwi-Bound piRNA Production by Distinct Mechanisms. Molecular cell 59, 819–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weick EM, and Miska EA (2014). piRNAs: from biogenesis to function. Development 141, 3458–3471. [DOI] [PubMed] [Google Scholar]
- Yang P, Wang Y, and Macfarlan TS (2017). The Role of KRAB-ZFPs in Transposable Element Repression and Mammalian Evolution. Trends in genetics : TIG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Xu J, Koppetsch BS, Wang J, Tipping C, Ma S, Weng Z, Theurkauf WE, and Zamore PD (2011). Heterotypic piRNA Ping-Pong requires qin, a protein with both E3 ligase and Tudor domains. Molecular cell 44, 572–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang J, Wang J, Theurkauf W, and Weng Z (2014). TEMP: a computational method for analyzing transposable element polymorphism in populations. Nucleic acids research 42, 6826–6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Hotspots for HMS-Beagle, blood, 3S18, and I-element. Related to Figure 4
Table S3. Summary of genomic libraries. Related to STAR methods
Figure S1. Monitoring transposition with an eGFP reporter indicates I-element preferentially inserts into oocyte genome, Related to Figure 1
(A) Schematic design of eGFP reporter to track I-element mobilization.
(B) Fly cross scheme to collect samples for measuring the potential transposition events from invaded and control flies.
(C) Probing transposition events by PCR. Transpositions in oocytes give rise to the PCR product without the intron. Primers are depicted in panel A. Samples were boxed with different color to match the corresponding materials depicted in panel B.
Figure S2. Upon activation, I-element mRNAs are enriched in oocytes in a microtubule-dependent manner, Related to Figure 1 and Figure 5
RNA-FISH to detect the I-element transcripts in invaded ovaries. The enrichment is abolished when the microtubule (M.T.)-mediated transport is blocked (Demecolcine treatment). Data are represented as mean ± SD. The egg chambers examined for each condition are from 9 animals.
Figure S3. Globally measure transposon mobilization in I-element-invaded progeny, Related to Figure 2
(A) Fly cross scheme.
(B) Scatter plot to display the detected potential new insertions.
Figure S4. Fly cross scheme to globally measure transposon mobilization upon Ago3&Aub depletion in ovaries, Related to Figure 3
(A) Fly cross scheme.
(B) Immuno-staining pictures to demonstrate the RNAi efficiency for Ago3 and Aub depletion.
Figure S5. Loss of Ago3&Aub leads to transposon insertions accumulated in oocytes, Related to Figure 3
(A) Genome sequencing to globally probe new transposition events from carcass samples, which serve as negative control.
(B) The new transposition events from oocytes were plotted to demonstrate both fold change and P-value for each transposon family. GLD = Germline Depletion.
(C) The number of detected new insertions shows linear correlation with the fold of genome coverage.
Figure S6. Blocking microtubule-mediated transport alleviates DNA damage in oocyte upon retrotransposon activation, Related to Figure 5
(A and B) Quantification of oocytes that do not have DNA damage signal upon piRNA pathway perturbation (A) or I-element invasion (B). GLD = Germline Depletion. Note, since we still count the oocytes that have weak γ-H2Av signals as DNA damage positive from Demecolcine-treated animals, we most likely have underestimated the rescue phenotype upon blocking microtubule transport. Data are represented as mean ± SD. The oocytes examined for each condition are from 9 animals.
(C) DAPI staining to measure karyosome morphology. Transposon mobilization in oocytes leads to karyosome packing defects that can be rescued by depolymerizing microtubule. In normal oocyte, the DNA is packed into a round condensed structure, named karyosome. Depleting Ago3 and Aub in germ cells results in karyosome packing defects (either stretched or fragmented). Because blocking microtubule-mediated transport made only 54% of karyosomes from control animals (White-depleted) are normal, depolymerizing microtubule thus appears to rescue the defects in Ago3&Aub depleted ovaries to control level (51%). GLD = Germline Depletion. Data are represented as mean ± SD. The oocytes examined for each condition are from 9 animals.
(D) Gurken staining to validate the effect of Demecolcine on microtubule-mediated transport.
Figure S7. Neither abundance nor localization of transposon mRNAs reflects mobility, Related to Figure 3 and Figure 7
(A) Scatter plots to display the number of new insertions detected in oocytes and the abundance of transposon mRNAs in ovaries. GLD = Germline Depletion.
(B) RNA-FISH to detect the localization of Burdock mRNA. Abundant Burdock mRNAs enrich in oocytes in the microtubule-dependent manner, but Burdock rarely mobilizes.
Data Availability Statement
All sequencing data generated from this study are available at NCBI: SRP144483.