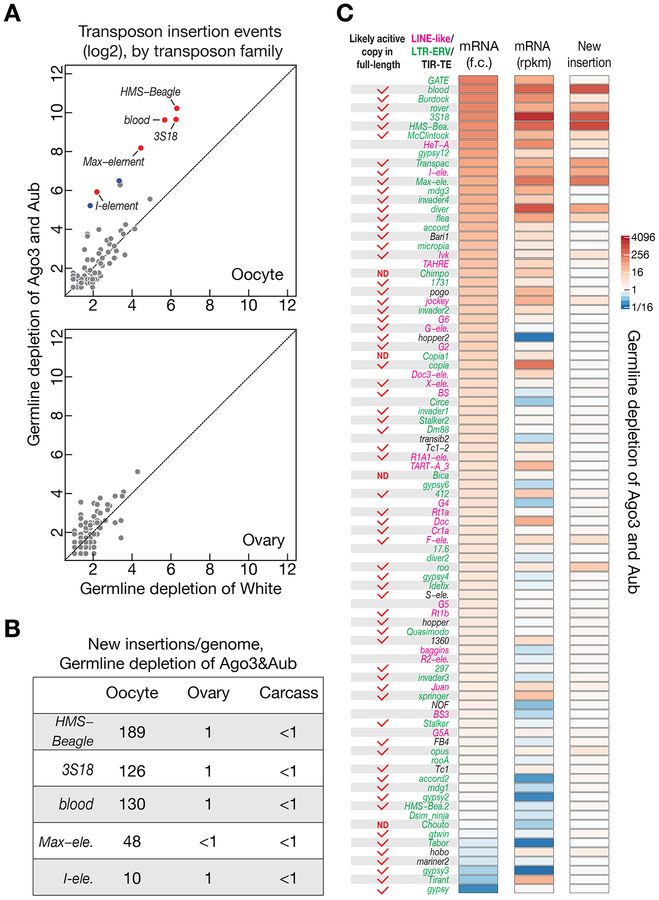
Figure 3. A group of retrotransposons preferentially mobilizes into oocyte genome.
(A) Genome sequencing to globally probe new transposition events in White (control) or Ago3&Aub depleted progeny. Upon piRNA pathway perturbation, retrotransposons preferentially insert into oocyte genome. Each dot represents one transposon family. Red: significantly changed for more than 10-fold. Blue: significantly changed for more than 5-fold but less than 10-fold.
(B) The number of new insertions (per genome) detected for the top 5 active transposons in Ago3&Aub depleted progeny. We found that the number of new insertions shows linear correlation with the number of genome coverage (Figure S5C).
(C) Upon piRNA perturbation, neither fold-change nor the absolute mRNA abundance in ovaries could faithfully reflect transposition activity in oocytes. RNA-Seq data and genome sequencing data were generated from the flies have identical genotypes. The change of mRNA abundance was quantified as fold change, and transposition events were calculated by subtracting the potential false positive numbers (from White depletion) from the insertion numbers detected in the oocytes laid by Ago3&Aub depleted progeny. The insertion events detected from 5× genome coverage were used to generate figures for panel A and C. ND = not determined yet.