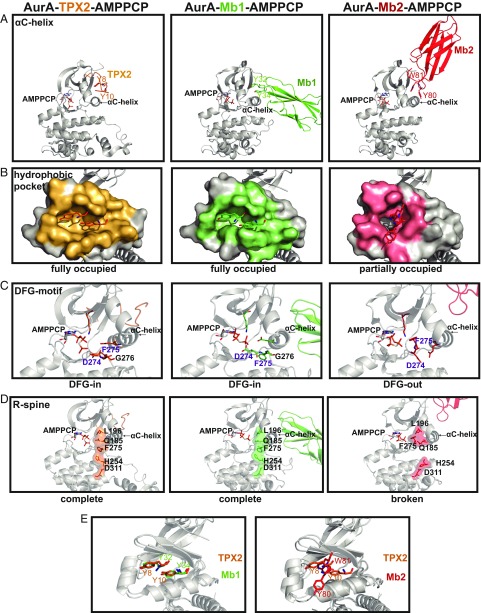
Fig. 5.
Monobodies stabilize either an active or inactive conformation of AurA. (A) Structures of AurA bound to TPX2 (PDB ID code 4C3P), Mb1 (PDB ID code 5G15), or Mb2 (PDB ID code 6C83) highlighting (A) differential engagement of AurA’s αC-helix by 2 aromatic residues within TPX2 or Mb1/Mb2, (B) AurA residues interfacing with TPX2 (orange), Mb1 (green) or Mb2 (pink) were calculated using PISA (40), (C) Hallmarks of an active kinase: DFG-in motif, K162-E181 salt bridge, and (D) completion of the R-spine are present in the AurA-TPX2 or AurA-Mb1 complexes and absent in the AurA-Mb2 structure. (E) Y32 and Y34 of Mb1 are superimposable with Y8 and Y10 of TPX2 (Left), whereas Y80 and W81 of Mb2 interact very differently with AurA (Right); see also SI Appendix, Fig. S5.