Abstract
MicroRNAs (miRNAs) are considered to be involved in the pathogenic initiation and progression of chronic nonbacterial prostatitis (CNP); however, the comprehensive expression profile of dysregulated miRNAs, relevant signaling pathways, and core machineries in CNP have not been fully elucidated. In the current research, CNP rat models were established through the intraprostatic injection of carrageenan into the prostate. Then, next-generation sequencing was performed to explore the miRNA expression profile in CNP. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) bioinformatical analyses were conducted to reveal the enriched biological processes, molecular functions, and cellular components and signaling pathways. As a result, 1224, 1039, and 1029 known miRNAs were annotated in prostate tissues from the blank control (BC), normal saline injection (NS), and carrageenan injection (CAR) groups (n = 3 for each group), respectively. Among them, 84 miRNAs (CAR vs BC) and 70 miRNAs (CAR vs NS) with significantly different expression levels were identified. Compared with previously reported miRNAs with altered expression in various inflammatory diseases, the majority of deregulated miRNAs in CNP, such as miR-146b-5p, miR-155-5p, miR-150-5p, and miR-139-5p, showed similar expression patterns. Moreover, bioinformatics analyses have enriched mitogen-activated protein kinase (MAPK), cyclic adenosine monophosphate (cAMP), endocytosis, mammalian target of rapamycin (mTOR), and forkhead box O (FoxO) signaling pathways. These pathways were all involved in immune response, which indicates the critical regulatory role of the immune system in CNP initiation and progression. Our investigation has presented a global view of the differentially expressed miRNAs and potential regulatory networks containing their target genes, which may be helpful for identifying the novel mechanisms of miRNAs in immune regulation and effective target-specific theragnosis for CNP.
Keywords: chronic nonbacterial prostatitis, immune, inflammation, miRNA, next-generation sequencing
INTRODUCTION
Inflammation is regarded as the driving factor in many diseases, including atherosclerosis, cancer, autoimmunity, and infections.1 It is broadly defined as a protective response by the organism in the presence of invaded pathogens or endogenous stress signals that subsequently results in the elimination of initial injury causes, necrotic cell clearance, and eventually facilitates tissue repair. Immunological processes are often involved in inflammatory responses.2 The pro-inflammatory cytokines and chemokines released by host cells such as tumor necrosis factor (TNF) and interleukin-1β (IL-1β) have autocrine and paracrine effects, leading to the local activation of macrophages and neutrophils. Meanwhile, cytokine-mediated activation of endothelial cells can increase vascular permeability and elicit the entrance of immune cells into tissues.3 Next, infiltrated type 1 helper T (TH1) cells secrete interferon-γ (IFN-γ) to activate neutrophils. Meanwhile, IL-17-producing helper T cells (TH17 cells) and innate lymphoid cells could secrete IL-22 to further stimulate the production and release of antimicrobial peptides.4 For instance, autoimmune diseases such as rheumatoid arthritis5 and inflammatory skin diseases6 caused by immunological imbalances have been frequently linked with chronic lesion inflammation. Likewise, pro-inflammatory cytokines can promote the progression of immune diseases by generating pathogenic B- and T-cells.7 Therefore, immune responses are important parts of inflammation.
Chronic nonbacterial prostatitis (CNP) is a commonly observed distress in male patients that is characterized by genital or pelvic discomfort, voiding and pain symptoms, and even by sexual dysfunction;8 it is much more common than chronic bacterial prostatitis (CBP), comprising more than 90% of chronic prostatitis.9 To date, the etiology of CNP remains largely unknown. During the past decades, several hypotheses have been proposed to explain CNP pathogenesis including defective urothelial integrity and function, cryptic infections, autoimmunity, endocrine imbalances, neuroplasticity, and psychosocial conditions.10 Recently, accumulating evidences from studies in patients and animal models have raised the possibility that CNP is a consequence of dysregulated inflammation associated with autoimmune disorders.11 Immunization with prostate gland homogenates or purified prostate proteins such as prostate–steroid binding protein and prostate acid phosphatase or peptides could induce prostatic inflammation and chronic pelvic pain.11 Consistently, T-cells with self-reactivity to prostate-specific antigen (PSA), prostatic acid phosphatase, and other prostatic and seminal plasma proteins have been successfully isolated from CNP patients.12
MicroRNAs (miRNAs) are posttranscriptional repressors of gene expression that play critical roles in a variety of physiological and pathological processes including development, differentiation, aging, and cancer. Notably, miRNAs are also widely accepted to participate in inflammation and lymphocyte homeostasis and allergic responses through coordinated modulation of multiple target genes. For example, miRNAs can execute the functional regulation of immune cells such as Th cells13 or inflammatory signal pathways such as the nuclear factor kappa-light-chain enhancer of activated B-cell (NF-κB) network14 in inflammatory or autoimmune diseases, indicating the crucial role of miRNAs in both inflammation and immune responses.
Recently, few studies have focused on the miRNA in CNP. One study revealed a decreased risk of erectile dysfunction (ED) in patients with CNP who were carriers of the miR-146a rs2910164 C allele,15 while another study found that N-acetylcysteine could ameliorate carrageenan-induced prostatitis and prostatic pain through miR-141-regulated Kelch-like ECH-associated protein-1/nuclear factor erythroid-2-related factor 2 (Keap1/Nrf2) signaling pathways.16 In 2018, a study evaluated the miRNA expression profile in expressed prostatic secretion (EPS).17 However, EPS is limited when illustrating the integral immune response and inflammation status of prostate tissue. Therefore, we comprehensively evaluated the dysregulated expression profile of miRNAs in CNP prostate samples using next-generation sequencing (NGS) technology, aiming to fully understand the role of miRNAs in CNP initiation and progression. Furthermore, bioinformatics analyses were applied to enrich immune-related biological processes, molecular functions, cellular components, and signaling pathways, the efforts of which would help explore the immune regulatory mechanisms in CNP.
MATERIALS AND METHODS
Experimental animals
Eighteen healthy adult male Wistar rats (aged 8 weeks, 250 ± 20 g) were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd., Beijing, China. The animals were housed in groups of three in plastic cages with soft bedding and free access to food and water under a 12/12 h reversed light-dark cycle. All animals were acclimated for 1 week before experimental procedures. All experimental protocols were approved by the Animal Research Ethical Committee of Anhui Medical University, Hefei, China. All surgeries were performed under chloral hydrate anesthesia, and all efforts were made to minimize suffering.
Carrageenan-induced CNP model establishment
Nine Wistar rats were randomly divided into the following three groups: (1) blank control (without any interference, BC) group, (2) normal saline injection (NS) group, and (3) carrageenan injection (CAR) group. For injection of carrageenan, rats were anesthetized with an intraperitoneal injection of 4% chloral hydrate (A600288; Sangon Biotech, Shanghai, China) and fixed in a supine position. Then, the lower abdomen above the rats' penis was shaved and the skin in this area was disinfected using three applications of 10% povidone-iodine solution (A606166; Sangon Biotech). A small midline incision was made in the disinfected area, and the bladder and prostate were carefully exposed. Using a microsyringe, 50 μl of sterile suspension of 5% carrageenan suspension (22049; Sigma-Aldrich, St. Louis, MO, USA) was injected into each ventral lobe in the prostate gland.18 A commensurate amount of sterile normal saline was injected in the NS group and nothing was injected in the BC group. After checking for bleeding at the dissection site, the wound was closed in layers.
Sample collection
One week later, all the rats were deprived of food and water for 12 h and then sacrificed under anesthesia with chloral hydrate. Prostate samples were collected and rinsed with phosphate-buffered saline (PBS) to remove adherent blood and fat tissues. One ventral lobe was formalin fixed for hematoxylin and eosin (H&E) and immunohistochemistry (IHC) staining and the other ventral lobe was cut into pieces, stored in RNAlater solution (AM7020; Ambion, Carlsbad, CA, USA), and frozen at −80°C according to a standard operation procedure.
Histological analyses
The rat prostate tissues were fixed in 10% formalin, dehydrated, and then embedded in paraffin. Sections about 4-μm thickness were sliced from each embedded tissue and stained with HE. For HE staining, the standard protocol was followed. Briefly, 4 μm-sized paraffin-embedded tissue slides were dried at 65°C for 2 h and de-paraffinized with xylene (Sinopharm Chemical Reagent, Shanghai, China) for four consecutive times (10 min each time). The sections were then dipped into 100%, 95%, 80%, and 70% ethanol for 5 min each time. The sections were rinsed with tap water for 2 min to remove ethanol and then stained with hematoxylin for 5 min, followed by tap water washing for 3 min. Subsequently, the slides were dipped into 0.1% HCl (Sinopharm Chemical Reagent) for three times and washed with tap water, followed by 50°C water incubation for 5 min. Later on, the slides were stained with eosin for 2 min and then washed with tap water for 2 min. Finally, after dehydration and dipping the slides into xylene, 2–3 drops of mountant were added onto the slide for observation under a microscope (IX 71; Olympus, Tokyo, Japan).
For cyclooxygenase-2 (COX-2) expression evaluation, IHC analysis was performed with COX-2 antibody (E303; Spring Bioscience, Pleasanton, CA, USA) on whole-tissue sections. Briefly, after routine deparaffinization and rehydration, the slides were treated with 10 mmol l−1 sodium citrate buffer (pH = 6.0; ZLI-9065; Zhongshan Golden Bridge Biotechnology Co., Ltd., Beijing, China) and boiled for 10 min to retrieve antigen. The endogenous peroxidase activity was quenched by 3% hydrogen peroxide (Sinopharm Chemical Reagent) for 15 min. After PBS (E607008; Sangon Biotech) washing for 15 min, the slides were blocked by bovine serum albumin (BSA, 37520; Thermo Fisher Scientific, Waltham, MA, USA) in a wet box and incubated for 10 min. BSA was then removed and the slides were incubated with primary COX-2 antibody with 1:100 dilutions overnight at 4°C. The next day, the incubated slides were washed with PBS for three times (5 min each time) and further incubated with poly-horseradish peroxidase (HRP) goat IgG (SP-9000; Zhongshan Golden Bridge Biotechnology Co., Ltd) for 15 min. After 15-min PBS washing (5 min each time), streptavidin/HRP was added to incubate for 15 min, and then washed with PBS for 15 min (5 min each time). Subsequently, detection was made using 3, 3′-diaminobenzidene as instructed (DAB kit, ZLI-9018; Zhongshan Golden Bridge Biotechnology Co., Ltd). Finally, the slides were counterstained with hematoxylin before microscopic observation. Cytoplasmic staining was scored as negative (-), weak (+), moderate (++), or strong (+++). A positive result required both cytoplasmic staining in >10% of cells and moderate-to-strong intensity, as previously described.19 Quantitative analysis of the brown positive staining was performed using Image-Pro Plus 6.0 (Media Cybernetics, Rockville, MD, USA).
Enzyme-linked immunosorbent assay (ELISA)
Serum IFN-γ concentrations were determined by using rat ELISA kit assay (E-EL-R0009C; Elabscience, Wuhan, China) according to the manufacturer's protocol. Briefly, 48-well plates were incubated with 100 μl standard solution or serum samples obtained from rats (three rats for each group) at 37°C for 90 min and the liquid was removed. After 100-μl biotinylated detection antibody incubation for 1 h at 37°C, each well was washed three times. Subsequently, 100-μl HRP conjugate was added and incubated at 37°C for 30 min, and then washed five times. Then, 90-μl substrate reagent was added and reacted at 37°C for 15 min. Finally, 50-μl stop solution was added. Absorbances were measured at 450 nm immediately. The sensitivity limit of the method was 18.75 pg ml−1.
RNA quality check, amplification, and NGS
Total RNA was collected from the three groups of tissues using the TRIzol reagent (15596-026; Life Technologies, Carlsbad, CA, USA). RNA quality check was assessed using an A260/A280 ratio (A: absorbance) between 1.8 and 2.2, an OD260/230 radio (OD: optical density) more than 2.0, a sample concentration >200 ng μl−1, and an RIN over 7 using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Then, ligation of 3'- and 5'-adapters was completed for each qualified RNA with T4 RNA Ligases (Thermo Fisher Scientific) with the presence of a phosphate group on the 5'-end and a hydroxyl group on the 3'-end of mature miRNAs. Next, reverse transcription (RT) was performed to make cDNA with the SuperScript IV revertase (Thermo Fisher Scientific), followed by PCR amplification. The acquired miRNA library was separated and purified by high-resolution polypropylene acyl ammonia gel electrophoresis (PAGE), and finally the miRNA library quality check was completed with a Qubit and an Agilent 2100 Bioanalyzer before NGS (Hiseq 2500; Illumina, San Diego, CA, USA).
NGS data analyses and novel miRNA identification
The miRNA sequencing data were further analyzed according to the methods described in a previous publication.20 Briefly, clean reads were acquired via the following steps: removing adaptor sequences, deleting sequences without 3'-adapters and inserting sequences, and omitting sequences with a Q20 ratio <60% and a length >18–36 bp using the FASTX-Toolkit software (http://hannonlab.cshl.edu/fastx_toolkit/). The qualified sequences were analyzed based on their length peak and underwent homogeneous BLAST alignment (http://blast.ncbi.nlm.nih.gov/) to mature miRNAs in miRBase (http://www.mirbase.org/),21 as well as reference genome alignment using Rfam (http://rfam.xfam.org/).22 Identification of known miRNAs was conducted by using the miRDeep2 tool (https://www.mdc-berlin.de/8551903/en/),23 which was also routinely used for novel miRNA exploration.
Bioinformatics analyses for differentially expressed miRNAs
The approaches described for the enrichment of functions and signaling pathways were frequently used in the present study. Briefly, target genes of differentially expressed miRNAs from three groups were predicted using RNAhybrid (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/)24 and miRanda (http://www.microrna.org/microrna/home.do)25 and analyzed with R package clusterProfiler (http://bioconductor.org/packages/release/bioc/html/clusterProfiler.html) before undergoing Gene Ontology (GO; http://www.geneontology.org) and Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.genome.jp/kegg/)26 enrichment analyses. After mapping the potential target genes to the dataset of GO annotations and KEGG pathway datasets, the enriched biological processes, molecular functions, cellular components, and signaling pathways were eventually sorted.
Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) verification for altered miRNA and gene expression
cDNA synthesis was performed using a FastQuant RT kit (with gDNAase) following the manufacturer's instructions (KR10; Tiangen Biotech, Beijing, China). The abundance of individual miRNA was detected via an Applied Biosystems 7500 PCR System (Applied Biosystems, Foster City, CA, USA) using SuperReal PreMix Plus (SYBR Green; FP215, Tiangen Biotech). The specific RT and qPCR primers for all miRNAs and genes were purchased from Sangon Biotech and listed in Supplementary Table 1. The qPCR procedures were performed according to the protocols provided by the manufacturers. Briefly, triplicate reactions were performed at 95°C for 15 min, and the subsequent 40 amplification cycles were conducted at 95°C for 10 s and 60°C for 32 s. Meanwhile, U6 snRNA or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served as an internal normalized control. Relative miRNA abundances were calculated using 2−ΔΔCt (threshold cycle) formula, where ΔCt = CtmiRNA/gene − CtU6 snRNA/GAPDH and ΔΔCt = ΔCtCAR − ΔCtBC/NS. The miRNA/gene abundance differences between CAR and BC or NS group were analyzed by using unpaired t-tests. P < 0.05 indicated a statistically significant difference.
Supplementary Table 1.
Primers used for quantitative reverse transcription-polymerase chain reaction
| Name | Primer for reverse transcription | Gene-specific primer for real-time PCR | Universal primer for real-time PCR |
|---|---|---|---|
| rno-miR-155-5p | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGACCCCTAT | ACACTCCAGCTGGGTTAATGCTAATTGTG | TGGTGTCGTGGAGTCG |
| rno-miR-212-5p | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCAGTAAGC | ACACTCCAGCTGGGACCTTGGCTCTAGACT | |
| rno-miR-223-5p | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCAACTCAG | ACACTCCAGCTGGGCGTGTATTTGACAAG | |
| rno-miR-340-5p | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAATCAGTC | ACACTCCAGCTGGGTTATAAAGCAATGA | |
| rno-miR-210-3p | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTCAGCCGC | ACACTCCAGCTGGGCTGTGCGTGTGACA | |
| rno-miR-33-5p | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTGCAATGC | ACACTCCAGCTGGGGTGCATTGTAGTT | |
| rno-miR-130a-3p | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGATGCCCTT | ACACTCCAGCTGGGCAGTGCAATGTTAA | |
| rno-miR-19b-3p | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTCAGTTTT | ACACTCCAGCTGGGTGTGCAAATCCATGC | |
| rno-miR-26a-3p | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGGTGCAAGT | ACACTCCAGCTGGGCCTATTCTTGGTT | |
| rno-miR-145-5p | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAGGGATTC | ACACTCCAGCTGGGGTCCAGTTTTCCCAG | |
| U6 snRNA | ACGAATTTGCGTGTCATCC | CAGCACATATACTAAAATTGGAACG | ACGAATTTGCGTGTCATCC |
| cAMP | Oligo dT | GACATCAGTTGTGATGCGCC | AAGGCAGGCCTACTGCTCTA |
| mTOR | GCAATGGGCACGAGTTTGTT | AGTGTGTTCACCAGGCCAAA | |
| GAPDH | AGTGCCAGCCTCGTCTCATA | GAGAAGGCAGCCCTGGTAAC |
PCR: polymerase chain reaction; GAPDH: glyceraldehyde-3-phosphate dehydrogenase; cAMP: cyclic adenosine monophosphate; mTOR: mammalian target of rapamycin
Statistical analysis
The differences among groups were analyzed using a randomized block design analysis of variance with SPSS version 10.0 software (SPSS Inc., Chicago, IL, USA). P < 0.05 was considered statistically significant.
RESULTS
Inflammatory cell accumulation and COX-2 expression in CNP tissues
Intraprostatic carrageenan injection induced prostatic edema and increased inflammatory cell accumulation. The majority of inflammatory cells were neutrophils and presented to the interstitial space in the CAR group, while no inflammatory cell infiltration was observed in the BC or NS group (Supplementary Figure 1 (1.1MB, tif) ). Consistently, COX-2 protein levels were significantly higher (P < 0.01) in the prostates from the CAR group compared with those of the BC and NS groups (Supplementary Figure 2 (612.2KB, tif) ). The HE and IHC staining results indicated that the CNP models were successfully established.
Serum IFN-γ levels were elevated in CNP rats
IFN-γ has been widely recognized as a signature pro-inflammatory cytokine that plays a central role in inflammation, and several studies have suggested a crucial role for IFN-γ in the pathogenesis of experimental prostatitis in animal models.27,28 Thus, we have evaluated the serum concentration of the pro-inflammatory cytokine in our carrageenan-induced prostatitis model. As a result, concentrations of IFN-γ in BC and NS groups were 24.41 ± 1.50 pg ml−1 and 19.55 ± 1.13 pg ml−1, respectively, while CAR group showed a significantly increased level of IFN-γ at 44.11 ± 1.22 pg ml−1 (mean ± standard error of the mean [s.e.m.]; CAR vs BC, P = 0.0005; CAR vs NS, P = 0.0001). The elevated serum levels verified that CAR injection indeed elicited CNP in rats (Supplementary Figure 3 (418.1KB, tif) ).
Overview of miRNA-NGS data from CNP prostate specimens
The majority of the clean sequencing reads were 22 nucleotides (nt) in size, followed by 21-nt or 23-nt RNA fragments, which were within the typical size range of registered miRNAs (Figure 1a–1c). The 18–36-nt qualified sequences were further analyzed based on their length peak. The counts of mature miRNAs aligned to miRBase for each sample were as follows: 626, 602, and 564 for the BC group; 243, 632, and 608 for the NS group; and 425, 453, and 525 for the CAR group. After comparing the sequences from each sample to the Rfam database, the miRNA sequence number and proportion of the genome for miRNAs are presented in Figure 1d–1f, revealing that the ratio of miRNAs in each sample was >85%.
Figure 1.
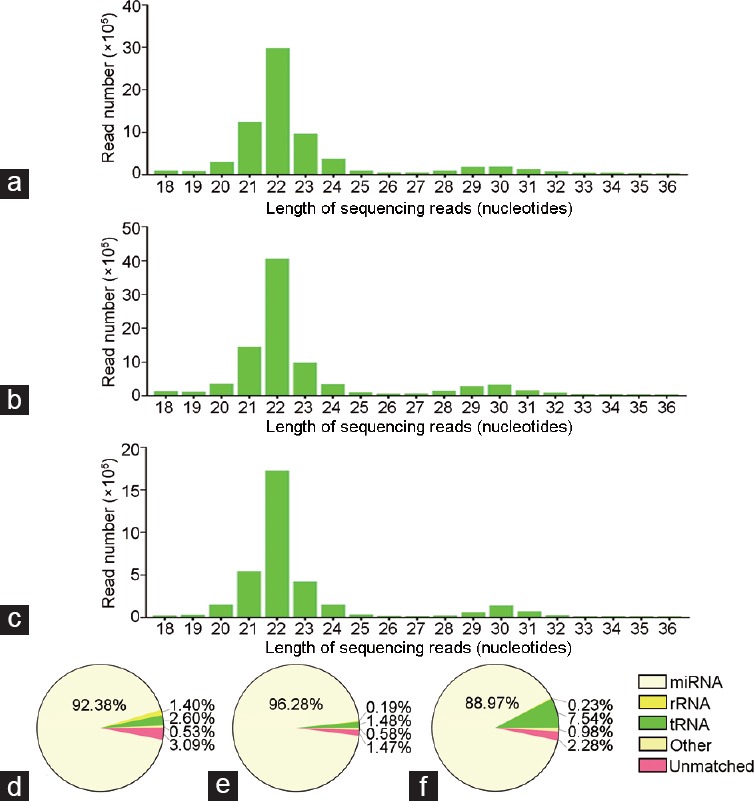
Length distribution of clean reads from small RNA NGS and Rfam alignment analyses in different sequencing samples. Length distribution (nucleotides) of clean reads from small RNA NGS of prostatic tissues (a) without any treatment (BC), (b) treated with NS, and (c) treated with CAR. The ratio of different smRNAs (miRNAs, rRNAs, scRNAs, snoRNAs, snRNAs, and tRNAs) compared with Rfam database from small RNA NGS of prostatic tissues (d) without any treatment (BC), (e) treated with NS, and (f) treated with CAR. NGS: next-generation sequencing; BC: blank control group; NS: normal saline group; CAR: carrageenan group.
Characteristics of top abundant known and novel miRNAs
To analyze the most abundant miRNAs in the prostate tissues, we listed the top twenty highly expressed miRNAs with relatively more read count in decreasing order from the known miRNAs in the CAR, NS, and BC groups, respectively, whose expression profiles were indicated by the average read counts. In the inflammatory prostate tissues, rno-miR-10a-5p, rno-miR-148a-3p, rno-miR-21-5p, rno-miR-10b-5p, rno-miR-143-3p, rno-miR-99a-5p, rno-miR-99b-5p, rno-miR-200b-3p, rno-miR-100-5p, and rno-miR-183-5p were the top ten abundant miRNAs. The highly expressed miRNAs in the NS group were rno-miR-148a-3p, rno-miR-10a-5p, rno-miR-200b-3p, rno-miR-143-3p, rno-miR-99a-5p, rno-miR-10b-5p, rno-miR-21-5p, rno-miR-100-5p, rno-miR-183-5p, and rno-miR-99b-5p. Correspondingly, the miRNAs possessing the highest expression levels in the BC group were rno-miR-148a-3p, rno-miR-200b-3p, rno-miR-10a-5p, rno-miR-143-3p, rno-miR-99a-5p, rno-miR-21-5p, rno-miR-10b-5p, rno-miR-152-3p, rno-miR-100-5p, and rno-miR-200c-3p. From these data, we found that the most abundantly expressed miRNAs in the prostate tissues from the three groups were approximately the same (Supplementary Table 2), indicating the identical histological origin of the paired samples undergoing NGS. Furthermore, 115 novel miRNAs were identified and presented in Supplementary Table 3, whose roles in the development and progression of prostatitis require further investigation.
Supplementary Table 2.
The top twenty most abundant known miRNAs expressed in prostate tissues from carrageenan group, normal saline group, and blank control group
| miRNA name |
BC group
Average count |
miRNA name |
NS group
Average count |
miRNA name |
CAR group
Average count |
|---|---|---|---|---|---|
| rno-miR-148a-3p | 1241921.667 | rno-miR-148a-3p | 1122445.667 | rno-miR-10a-5p | 715513.000 |
| rno-miR-200b-3p | 927917.000 | rno-miR-10a-5p | 852980.333 | rno-miR-148a-3p | 406646.333 |
| rno-miR-10a-5p | 690299.667 | rno-miR-200b-3p | 489547.667 | rno-miR-21-5p | 341285.667 |
| rno-miR-143-3p | 525922.333 | rno-miR-143-3p | 352674.667 | rno-miR-10b-5p | 197378.667 |
| rno-miR-99a-5p | 299589.333 | rno-miR-99a-5p | 223992.667 | rno-miR-143-3p | 164819.333 |
| rno-miR-21-5p | 293555.667 | rno-miR-10b-5p | 205457.667 | rno-miR-99a-5p | 141099.333 |
| rno-miR-10b-5p | 177072.667 | rno-miR-21-5p | 169482.333 | rno-miR-99b-5p | 100704.667 |
| rno-miR-152-3p | 101867.667 | rno-miR-100-5p | 117617.333 | rno-miR-200b-3p | 98399.667 |
| rno-miR-100-5p | 85266.000 | rno-miR-183-5p | 94032.667 | rno-miR-100-5p | 85978.000 |
| rno-miR-200c-3p | 68235.667 | rno-miR-99b-5p | 64474.000 | rno-miR-183-5p | 59241.333 |
| rno-miR-125b-5p | 67222.333 | rno-miR-152-3p | 62699.333 | rno-miR-152-3p | 46765.333 |
| rno-miR-30d-5p | 51155.000 | rno-miR-125b-5p | 43171.000 | rno-miR-182 | 23234.333 |
| rno-miR-126a-3p | 49437.667 | rno-miR-30d-5p | 36698.000 | rno-miR-30d-5p | 20983.000 |
| rno-miR-183-5p | 49143.667 | rno-miR-200c-3p | 33285.000 | rno-let-7f-5p | 20961.000 |
| rno-miR-99b-5p | 44799.333 | rno-miR-182 | 32054.333 | rno-miR-148b-3p | 13573.000 |
| rno-miR-200a-3p | 42278.667 | rno-miR-148b-3p | 24688.667 | rno-miR-125b-5p | 12261.000 |
| rno-miR-182 | 38630.667 | rno-miR-126a-3p | 20247.000 | rno-miR-101a-3p | 10525.000 |
| rno-let-7f-5p | 25440.667 | rno-miR-200a-3p | 19071.333 | rno-miR-126a-3p | 10474.000 |
| rno-miR-148b-3p | 25121.000 | rno-let-7f-5p | 18518.333 | rno-miR-200c-3p | 8360.000 |
| rno-miR-101a-3p | 20355.333 | rno-miR-342-3p | 17012.333 | rno-miR-146a-5p | 7374.333 |
The top abundant miRNAs overlapped in all the three groups are emphasized in bold fonts. CAR: carrageenan group; BC: blank control group; NS: normal saline group
Supplementary Table 3.
The novel miRNAs identified by next-generation sequencing in prostate tissues from carrageenan group, normal saline group, and blank control groups
| miRNA | BC | NS | CAR |
|---|---|---|---|
| 1_72 | 0.3 | 0.7 | 2.0 |
| 2_10536 | 1.0 | 1.3 | 0.3 |
| 5_16527 | 12.3 | 7.3 | 0.0 |
| 8_19321 | 55.7 | 10.0 | 25.0 |
| 10_4026 | 3.3 | 1.3 | 0.7 |
| 9_20141 | 1.3 | 0.0 | 1.7 |
| 4_14002 | 3.7 | 2.7 | 3.0 |
| 15_7332 | 6.3 | 7.0 | 0.3 |
| 15_7495 | 0.0 | 0.0 | 6.7 |
| 4_15090 | 1.7 | 1.3 | 0.0 |
| 7_18253 | 0.7 | 0.3 | 0.3 |
| 10_2857 | 39.3 | 22.7 | 39.0 |
| 1_829 | 1.3 | 0.7 | 2.0 |
| 7_18655 | 0.0 | 3.0 | 0.0 |
| 1_1633 | 3.3 | 0.7 | 0.3 |
| 10_2671 | 1.0 | 1.3 | 1.7 |
| 9_20621 | 37.3 | 31.7 | 4.3 |
| 1_25 | 2.0 | 1.3 | 1.3 |
| 9_20250 | 1.7 | 1.3 | 0.7 |
| 3_13516 | 2.0 | 2.3 | 5.3 |
| 7_17854 | 35.3 | 40.0 | 22.3 |
| 9_20741 | 2.0 | 2.3 | 0.0 |
| 13_5819 | 1.0 | 2.3 | 0.7 |
| 7_17969 | 42.3 | 39.3 | 197.3 |
| 15_7174 | 753.0 | 97.7 | 3.7 |
| 7_17853 | 33.3 | 38.0 | 21.0 |
| 1_1023 | 2.0 | 2.7 | 3.0 |
| 17_8769 | 21.3 | 21.3 | 38.3 |
| 1_1074 | 0.7 | 0.3 | 0.0 |
| 6_17598 | 39.3 | 22.7 | 39.0 |
| 2_10987 | 1.0 | 0.7 | 1.0 |
| 7_18649 | 0.0 | 3.0 | 0.0 |
| 11_4601 | 1.3 | 0.3 | 4.7 |
| 18_9644 | 49.7 | 22.7 | 43.3 |
| 14_6423 | 2.0 | 1.3 | 1.3 |
| 1_1131 | 93.0 | 16.0 | 0.0 |
| X_21112 | 6.0 | 4.7 | 1.0 |
| 2_11769 | 1.7 | 0.3 | 1.0 |
| 3_12581 | 59.7 | 52.3 | 5.0 |
| 15_7578 | 15.3 | 8.0 | 14.3 |
| 17_9029 | 1.0 | 1.0 | 4.7 |
| 6_17186 | 30.3 | 27.0 | 5.0 |
| 2_11409 | 1.7 | 3.0 | 0.0 |
| 9_20554 | 1.7 | 2.0 | 3.0 |
| 11_4711 | 0.3 | 1.0 | 0.0 |
| 4_14169 | 4.0 | 3.0 | 0.0 |
| 8_19379 | 9.7 | 7.3 | 0.0 |
| 7_18537 | 0.3 | 1.0 | 0.7 |
| 8_19665 | 0.0 | 0.0 | 1.3 |
| 8_19380 | 9.7 | 7.7 | 0.0 |
| 17_8821 | 4.3 | 5.0 | 2.0 |
| 2_11240 | 182.3 | 0.0 | 0.0 |
| 17_9040 | 3.3 | 4.3 | 30.3 |
| 1_1371 | 12.0 | 15.0 | 4.7 |
| 4_15026 | 39.3 | 22.7 | 39.0 |
| 3_13398 | 51.7 | 0.0 | 0.0 |
| 17_9036 | 3.3 | 4.3 | 30.3 |
| 10_3377 | 1.3 | 0.7 | 1.0 |
| 4_14852 | 20.0 | 12.7 | 1.7 |
| 10_3118 | 1.3 | 12.0 | 0.7 |
| 8_19062 | 0.0 | 3.0 | 0.0 |
| 11_4695 | 0.0 | 0.0 | 1.0 |
| 8_19221 | 44.7 | 0.0 | 0.0 |
| 4_14454 | 41.3 | 54.3 | 25.3 |
| 2_10926 | 4.7 | 0.0 | 0.0 |
| 13_5794 | 16.0 | 12.3 | 6.0 |
| 14_6594 | 733.7 | 137.3 | 0.0 |
| 17_8773 | 183.3 | 406.7 | 349.3 |
| 16_7829 | 0.0 | 1.0 | 0.0 |
| 1_2103 | 7.3 | 4.3 | 7.0 |
| 14_6599 | 5.7 | 8.3 | 1.7 |
| 3_13532 | 0.0 | 0.0 | 6.3 |
| 14_6985 | 3.7 | 2.7 | 3.0 |
| 8_19615 | 74.7 | 53.0 | 2.0 |
| X_21119 | 12.7 | 2.0 | 0.0 |
| 1_1372 | 12.0 | 15.0 | 4.7 |
| 1_66 | 0.0 | 1.0 | 0.0 |
| 20_12145 | 5.0 | 0.7 | 1.3 |
| 19_9750 | 96.0 | 0.0 | 0.0 |
| 7_18588 | 11.3 | 9.7 | 1.7 |
| 17_9038 | 3.3 | 4.3 | 30.3 |
| 1_2214 | 205.7 | 0.3 | 0.0 |
| 6_17033 | 3.7 | 3.0 | 4.3 |
| 17_8585 | 7.7 | 4.3 | 0.3 |
| 3_13730 | 1.7 | 1.7 | 0.0 |
| 10_3082 | 25.3 | 20.3 | 38.7 |
| 5_15288 | 355.3 | 111.7 | 45.3 |
| 20_12339 | 7.0 | 37.3 | 0.7 |
| X_21303 | 19.0 | 12.7 | 1.7 |
| 1_241 | 24.3 | 19.0 | 2.3 |
| 20_12255 | 5.0 | 0.7 | 1.3 |
| 13_6099 | 5.7 | 5.0 | 2.0 |
| 4_14453 | 41.3 | 54.3 | 25.3 |
| 1_1444 | 1.3 | 1.3 | 1.7 |
| 19_10062 | 22.0 | 0.0 | 0.0 |
| 7_18067 | 1.7 | 0.7 | 0.0 |
| 7_18607 | 1.7 | 1.7 | 0.7 |
| 7_18093 | 0.3 | 1.0 | 2.7 |
| 13_5579 | 5.3 | 0.0 | 0.0 |
| 17_9042 | 3.3 | 4.3 | 30.3 |
| 3_13728 | 1.7 | 1.7 | 0.0 |
| 1_2228 | 12.7 | 6.3 | 1.7 |
| 14_6676 | 1.3 | 0.3 | 0.0 |
| 5_15617 | 1.0 | 0.0 | 0.0 |
| 14_6534 | 22.7 | 14.3 | 1.0 |
| 4_13981 | 0.3 | 0.3 | 1.3 |
| 7_18089 | 296.3 | 0.0 | 0.0 |
| 6_17529 | 2.0 | 3.7 | 9.3 |
| 10_3621 | 152.0 | 75.7 | 5.3 |
| 12_5301 | 2.3 | 0.7 | 0.0 |
| 4_15030 | 31.7 | 9.0 | 0.0 |
| 1_2445 | 0.7 | 0.7 | 2.3 |
| 1_692 | 1.3 | 0.7 | 1.3 |
| 4_13943 | 2.0 | 0.3 | 1.3 |
| 9_20092 | 1.3 | 2.3 | 0.3 |
BC: blank control group; NS: normal saline group; CAR: carrageenan group
Differentially expressed profile of miRNAs in CNP tissues
When comparing the distinct levels of miRNA abundance among the three groups, two quantities were of great importance: one is the P value and another is the log2 (fold change) value. The P value responses to whether the difference in expression levels was statistically significant, while the log2 (fold change) value could illustrate the magnitude of the miRNA abundance differences. A log2 (fold change) value above 0 indicates upregulation and a log2 (fold change) value below 0 indicates decreased expression. Notably, even very large log2 (fold change) values may have insignificant P values caused by considerable variabilities. Similarly, relatively small magnitude in the differences may also possess tiny P values due to the highly consistent technical replicates within each sample. Thus, a useful visualization method called a “volcano plot,” which can simultaneously exhibit log2 (fold change) and P values on the X- and Y-axes, respectively, has been introduced to conduct our analysis. Generally, points located in the upper-left or upper-right regions of the plot and indicated with green or red color possess both small P values (<0.05) and high log2 (fold change). In this comparative study, the volcano plot generated by the miRNA profiles from the three groups is shown in Figure 2a and 2b. As a result, 84 (CAR vs BC) and 70 (CAR vs NS) significantly dysregulated known miRNAs were found after adjusting for multiple testing. Among them, the top twenty upregulated and downregulated miRNAs are shown in Table 1 (CAR vs BC) and Table 2 (CAR vs NS), respectively. Meanwhile, hierarchical clustering analysis was performed to cluster the same or similar differentially expressed miRNAs (Figure 2c and 2d).
Figure 2.
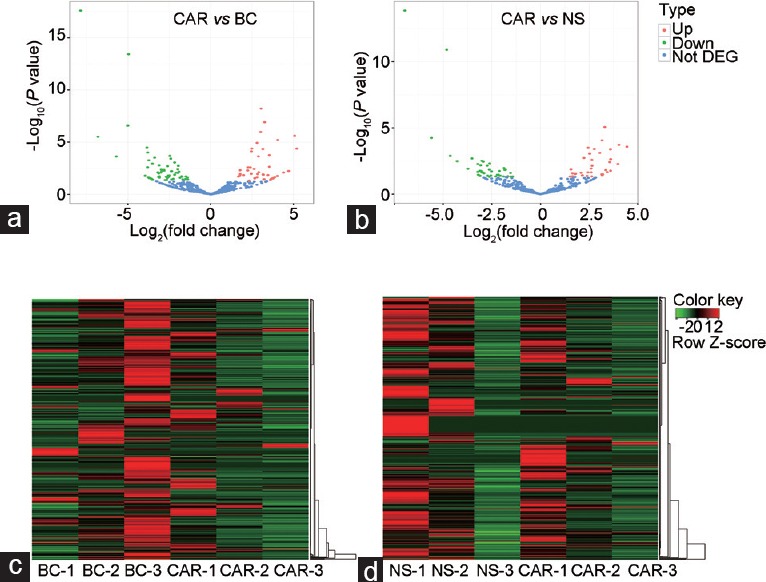
The overview of the volcano plot generated by miRNA profile and clustering heat maps of differentially expressed miRNA. The volcano plot generated by miRNA expression profile from small RNA NGS between prostatic tissues (a) treated with CAR or without any treatment (BC) and (b) treated with CAR or NS. The red points in the plot represent significantly upregulated miRNAs and the green points represent significantly downregulated miRNAs, while the blue points show no statistically significant differences due to either small fold change (<2) or insignificant P values. The heat map of miRNA expression profile between prostatic tissues (c) treated with CAR or without any treatment (BC) and (d) treated with CAR or NS. For c and d, each column represents a certain sample and lines represent different genes; the corresponding color represents the amount of gene expression in the sample: red color indicates higher expression and green represents lower expression. DEG: differentially expressed genes; BC: blank control group; NS: normal saline group; CAR: carrageenan group.
Table 1.
A collection of top twenty upregulated and downregulated miRNAs detected by next-generation sequencing in prostate tissues (carrageenan group vs blank control group)
| miRNA | BaseMean | Log2(fold change) | P | Type |
|---|---|---|---|---|
| rno-miR-155-5p* | 7.741 | 5.205 | <0.001 | Up |
| rno-miR-501-3p* | 2186.460 | 5.076 | <0.001 | Up |
| rno-miR-1956-3p* | 3.715 | 4.710 | 0.006 | Up |
| rno-miR-212-5p* | 109.202 | 4.057 | <0.001 | Up |
| rno-miR-146a-3p* | 14.179 | 3.554 | <0.001 | Up |
| rno-miR-223-5p* | 4.443 | 3.550 | 0.002 | Up |
| rno-miR-370-3p | 2.897 | 3.457 | 0.014 | Up |
| rno-miR-340-5p* | 326.544 | 3.265 | <0.001 | Up |
| rno-miR-6215* | 2.621 | 3.233 | 0.0362 | Up |
| rno-miR-511-3p* | 3.968 | 3.181 | 0.013 | Up |
| rno-miR-184* | 192.557 | 3.165 | 0.013 | Up |
| rno-miR-210-3p* | 85.837 | 3.155 | <0.001 | Up |
| rno-miR-146b-5p* | 2201.022 | 3.053 | <0.001 | Up |
| rno-miR-542-3p | 125.935 | 3.027 | <0.001 | Up |
| rno-miR-351-3p | 44.609 | 2.910 | 0.008 | Up |
| rno-miR-511-5p* | 20.242 | 2.888 | <0.001 | Up |
| rno-miR-382-5p | 7.463 | 2.843 | 0.012 | Up |
| rno-miR-450b-3p | 4.201 | 2.813 | 0.023 | Up |
| rno-miR-146a-5p* | 4459.610 | 2.810 | <0.001 | Up |
| rno-miR-541-5p | 109.988 | 2.627 | 0.004 | Up |
| rno-miR-33-5p* | 365.461 | −7.861 | <0.001 | Down |
| rno-miR-136-5p* | 20.234 | −6.805 | <0.001 | Down |
| rno-miR-19a-3p* | 8.844 | −5.697 | <0.001 | Down |
| rno-miR-150-5p | 1275.055 | −5.005 | <0.001 | Down |
| rno-miR-130a-3p* | 774.472 | −4.961 | <0.001 | Down |
| rno-miR-883-5p* | 25.518 | −3.852 | <0.001 | Down |
| rno-miR-874-5p* | 4.954 | −3.809 | 0.003 | Down |
| rno-miR-324-5p | 24.101 | −3.802 | <0.001 | Down |
| rno-miR-542-5p* | 12.549 | −3.738 | <0.001 | Down |
| rno-miR-19b-3p* | 45.434 | −3.517 | 0.001 | Down |
| rno-miR-1247-5p | 15.468 | −3.384 | <0.001 | Down |
| rno-miR-301a-3p | 11.139 | −3.190 | 0.008 | Down |
| rno-miR-547-5p* | 2.531 | −3.145 | 0.021 | Down |
| rno-miR-186-3p* | 4.322 | −3.139 | 0.010 | Down |
| rno-let-7i-3p* | 10.928 | −3.118 | 0.002 | Down |
| rno-miR-26a-3p* | 3.129 | −3.105 | 0.019 | Down |
| rno-miR-196b-3p* | 2.454 | −3.099 | 0.025 | Down |
| rno-miR-141-3p | 66.361 | −2.998 | 0.008 | Down |
| rno-miR-211-5p | 5.806 | −2.982 | 0.004 | Down |
| rno-miR-141-5p* | 11.748 | −2.957 | 0.006 | Down |
*The differentially expressed miRNAs overlapped in CAR versus BC and CAR versus NS. BaseMean: the average levels of miRNA expression; log2(fold change): the 2-logarithm value of the expression fold change of certain miRNA; NS: normal saline group; CAR: carrageenan group; BC: blank control group
Table 2.
A collection of top twenty upregulated and downregulated miRNAs detected by next-generation sequencing in prostate tissues (carrageenan group vs normal saline group)
| miRNA | BaseMean | Log2(fold change) | P | Type |
|---|---|---|---|---|
| rno-miR-155-5p* | 4.975 | 4.445 | <0.001 | Up |
| rno-miR-184* | 124.084 | 4.036 | <0.001 | Up |
| rno-miR-132-3p | 1.673 | 3.986 | 0.005 | Up |
| rno-miR-501-3p* | 1569.684 | 3.610 | 0.002 | Up |
| rno-miR-1956-3p* | 2.649 | 3.607 | 0.017 | Up |
| rno-miR-212-5p* | 74.382 | 3.600 | <0.001 | Up |
| rno-miR-223-5p* | 2.958 | 3.566 | 0.004 | Up |
| rno-miR-6215* | 1.753 | 3.435 | 0.022 | Up |
| rno-miR-6315 | 3.317 | 3.362 | 0.015 | Up |
| rno-miR-340-5p* | 215.995 | 3.313 | <0.001 | Up |
| rno-miR-203b-3p | 0.906 | 3.201 | 0.032 | Up |
| rno-miR-18a-5p | 1.114 | 3.180 | 0.037 | Up |
| rno-miR-146a-5p* | 2835.543 | 3.171 | <0.001 | Up |
| rno-miR-210-3p* | 56.777 | 3.048 | <0.001 | Up |
| rno-miR-146b-5p* | 1479.402 | 2.770 | <0.001 | Up |
| rno-miR-146a-3p* | 9.853 | 2.650 | 0.007 | Up |
| rno-miR-511-5p* | 13.219 | 2.643 | 0.003 | Up |
| rno-miR-511-3p* | 2.567 | 2.637 | 0.046 | Up |
| rno-miR-494-3p | 4.039 | 2.608 | 0.023 | Up |
| rno-miR-99b-3p | 4.444 | 2.605 | 0.012 | Up |
| rno-miR-33-5p* | 141.949 | −6.971 | <0.001 | Down |
| rno-miR-136-5p* | 9.496 | −5.599 | <0.001 | Down |
| rno-miR-130a-3p* | 492.946 | −4.816 | <0.001 | Down |
| rno-miR-19a-3p* | 2.963 | −4.633 | 0.001 | Down |
| rno-let-7g-3p | 3.414 | −4.275 | 0.003 | Down |
| rno-miR-190b-5p | 1.018 | −3.722 | 0.012 | Down |
| rno-miR-542-5p* | 7.249 | −3.517 | 0.002 | Down |
| rno-miR-141-5p* | 11.527 | −3.213 | 0.005 | Down |
| rno-miR-25-5p | 8.962 | −3.213 | 0.021 | Down |
| rno-miR-186-3p* | 2.877 | −3.099 | 0.017 | Down |
| rno-miR-19b-3p* | 24.007 | −3.073 | 0.007 | Down |
| rno-miR-124-3p | 13.443 | −3.036 | 0.018 | Down |
| rno-miR-133a-5p | 1.460 | −3.010 | 0.023 | Down |
| rno-let-7i-3p* | 6.655 | −3.005 | 0.003 | Down |
| rno-miR-26a-3p* | 1.714 | −2.959 | 0.023 | Down |
| rno-miR-7a-1-3p | 27.551 | −2.933 | 0.004 | Down |
| rno-miR-874-5p* | 1.678 | −2.900 | 0.026 | Down |
| rno-miR-883-5p* | 7.659 | −2.739 | 0.007 | Down |
| rno-miR-547-5p* | 1.101 | −2.717 | 0.040 | Down |
| rno-miR-196b-3p* | 1.004 | −2.629 | 0.048 | Down |
*The differentially expressed miRNAs overlapped in CAR versus BC and CAR versus NS. BaseMean: the average levels of miRNA expression; log2(fold change): the 2-logarithm value of the expression fold change of certain miRNA; CAR: carrageenan group; NS: normal saline group
As expected, the twenty upregulated and downregulated miRNAs between the CAR versus BC groups and the CAR versus NS groups mostly overlapped (Table 1 and 2), verifying the significant regulatory roles of miRNAs in the progression of inflammation in CNP. Meanwhile, the normalized expression levels of aforementioned top differentially expressed miRNAs in BC, NS, and CAR groups are shown in Supplementary Table 4. The most abundantly upregulated miRNAs in the prostate tissues were rno-miR-155-5p, rno-miR-501-3p, rno-miR-1956-3p, rno-miR-212-5p, and rno-miR-146a-3p for CAR versus BC and rno-miR-155-5p, rno-miR-184, rno-miR-132-3p, rno-miR-501-3p, and rno-miR-1956-3p for CAR versus NS. Meanwhile, rno-miR-33-5p, rno-miR-136-5p, rno-miR-19a-3p, rno-miR-150-5p, and rno-miR-130a-3p were the top five most abundantly downregulated miRNAs in the prostate tissues for CAR versus BC, while rno-miR-33-5p, rno-miR-136-5p, rno-miR-130a-3p, rno-miR-19a-3p, and rno-let-7g-3p were the five most decreased miRNAs for CAR compared with NS. Besides, we have compared the differential expression between BC and NS groups, the results showed almost no significant difference of the miRNA expression levels between the two groups except for rno-miR-124-3p and rno-miR-150-5p, indicating that the altered expression between CAR versus BC and CAR versus NS was reliable (Supplementary Table 5 and 6).
Supplementary Table 4.
The normalized expression levels of top differentially expressed miRNAs in blank control group, normal saline group, and carrageenan group
| miRNA | Read count for differential expression comparison (mean±s.d.) | ||
|---|---|---|---|
| BC | NS | CAR | |
| rno-let-7g-3p | 1.382±1.329 | 9.185±9.116 | 0 |
| rno-let-7i-3p | 16.807±7.080 | 16.563±6.086 | 1.330±1.475 |
| rno-miR-124-3p | 1.711±1.276 | 34.486±53.508 | 1.882±1.765 |
| rno-miR-1247-5p | 24.276±12.891 | 9.415±9.353 | 2.031±0.847 |
| rno-miR-130a-3p | 1238.82±423.456 | 1293.674±618.992 | 36.495±6.535 |
| rno-miR-132-3p | 1.713±2.236 | 0 | 4.418±4.300 |
| rno-miR-133a-5p | 2.459±0.612 | 3.755±4.066 | 0.194±0.337 |
| rno-miR-136-5p | 33.580±8.957 | 25.574±24.332 | 0 |
| rno-miR-141-3p | 101.783±60.506 | 63.841±78.036 | 10.017±14.649 |
| rno-miR-141-5p | 18.269±4.403 | 29.617±24.506 | 1.555±2.694 |
| rno-miR-146a-3p | 1.327±1.216 | 2.162±2.580 | 23.154±11.554 |
| rno-miR-146a-5p | 898.044±469.504 | 652.855±418.807 | 6766.379±1381.53 |
| rno-miR-146b-5p | 381.639±68.557 | 449.383±274.756 | 3400.372±639.633 |
| rno-miR-150-5p | 2180.166±2524.84 | 413.195±350.110 | 50.092±8.907 |
| rno-miR-155-5p | 0.177±0.306 | 0.152±0.263 | 12.929±6.725 |
| rno-miR-184 | 23.981±22.927 | 12.100±7.067 | 298.736±241.014 |
| rno-miR-186-3p | 6.763±3.118 | 7.425±10.220 | 0.389±0.673 |
| rno-miR-18a-5p | 1.510±1.049 | 0 | 3.111±5.388 |
| rno-miR-190b-5p | 2.233±1.967 | 2.711±2.998 | 0 |
| rno-miR-1956-3p | 0 | 0 | 6.460±10.687 |
| rno-miR-196b-3p | 3.812±2.390 | 2.499±2.312 | 0.194±0.337 |
| rno-miR-19a-3p | 15.237±11.641 | 7.887±9.065 | 0 |
| rno-miR-19b-3p | 72.224±27.199 | 59.459±43.900 | 4.783±6.109 |
| rno-miR-203b-3p | 1.349±1.514 | 0 | 2.342±3.563 |
| rno-miR-210-3p | 12.736±6.485 | 15.770±12.967 | 134.342±56.746 |
| rno-miR-211-5p | 9.031±2.253 | 2.347±2.115 | 0.747±0.648 |
| rno-miR-212-5p | 8.904±5.220 | 9.679±10.771 | 176.908±138.592 |
| rno-miR-223-5p | 0.299±0.517 | 0.245±0.424 | 7.283±3.123 |
| rno-miR-25-5p | 3.366±4.401 | 23.489±37.992 | 0.778±1.347 |
| rno-miR-26a-3p | 4.947±3.069 | 4.205±4.109 | 0.358±0.620 |
| rno-miR-301a-3p | 17.919±11.858 | 6.208±8.914 | 1.167±2.020 |
| rno-miR-324-5p | 39.445±26.288 | 14.142±12.991 | 1.882±1.765 |
| rno-miR-33-5p | 600.211±186.068 | 380.399±111.588 | 1.493±1.297 |
| rno-miR-340-5p | 48.069±5.015 | 44.795±17.153 | 516.615±196.374 |
| rno-miR-351-3p | 7.188±3.162 | 30.164±27.187 | 69.534±93.433 |
| rno-miR-370-3p | 0.352±0.609 | 0.304±0.527 | 4.49±3.549 |
| rno-miR-382-5p | 1.083±1.391 | 6.295±5.073 | 11.547±9.138 |
| rno-miR-450b-3p | 0.498±0.862 | 0.641±0.751 | 6.684±5.920 |
| rno-miR-494-3p | 4.720±5.967 | 0.793±0.741 | 9.369±6.893 |
| rno-miR-501-3p | 73.180±43.520 | 198.701±280.557 | 3685.298±4622.082 |
| rno-miR-511-3p | 0.551±0.529 | 0.489±0.847 | 6.502±8.048 |
| rno-miR-511-5p | 3.965±2.965 | 3.266±3.303 | 30.794±8.324 |
| rno-miR-541-5p | 23.101±6.752 | 31.035±31.754 | 164.612±159.34 |
| rno-miR-542-3p | 21.395±7.013 | 35.654±27.461 | 194.759±39.334 |
| rno-miR-542-5p | 19.730±11.624 | 18.585±17.694 | 0.941±0.882 |
| rno-miR-547-5p | 4.052±2.496 | 2.777±2.594 | 0.194±0.337 |
| rno-miR-6215 | 0.177±0.306 | 0 | 4.296±5.683 |
| rno-miR-6315 | 0.884±1.531 | 0.245±0.424 | 8.247±11.501 |
| rno-miR-7a-1-3p | 28.644±21.314 | 68.324±33.957 | 6.129±7.990 |
| rno-miR-874-5p | 7.834±1.536 | 4.152±4.425 | 0.358±0.620 |
| rno-miR-883-5p | 40.454±5.575 | 18.698±7.875 | 1.913±2.444 |
| rno-miR-99b-3p | 4.632±2.640 | 0.945±0.820 | 10.465±5.392 |
CAR: carrageenan group; BC: blank control group; NS: normal saline group; s.d.: standard deviation
Supplementary Table 5.
A collection of top twenty upregulated miRNAs detected by next-generation sequencing in prostate tissues (blank control group vs normal saline group)
| miRNA | baseMean | log2 (fold change) | P | Type |
|---|---|---|---|---|
| rno-miR-23b-5p | 7.402 | 3.501 | 0.271 | Not DEG |
| rno-miR-132-3p | 0.972 | 2.628 | 0.315 | Not DEG |
| rno-miR-150-5p | 1504.514 | 2.477 | 0.025 | Up |
| rno-miR-18a-5p | 0.883 | 2.429 | 0.335 | Not DEG |
| rno-miR-203b-3p | 0.721 | 2.223 | 0.437 | Not DEG |
| rno-miR-130b-5p | 1.339 | 2.124 | 0.298 | Not DEG |
| rno-miR-494-3p | 3.228 | 1.896 | 0.223 | Not DEG |
| rno-miR-423-5p | 286.706 | 1.768 | 0.065 | Not DEG |
| rno-miR-10a-3p | 22.937 | 1.728 | 0.116 | Not DEG |
| rno-miR-142-3p | 34.767 | 1.722 | 0.068 | Not DEG |
| rno-miR-140-5p | 10.152 | 1.721 | 0.188 | Not DEG |
| rno-miR-455-3p | 7.335 | 1.709 | 0.126 | Not DEG |
| rno-miR-883-3p | 30.651 | 1.694 | 0.068 | Not DEG |
| rno-let-7d-5p | 428.488 | 1.682 | 0.077 | Not DEG |
| rno-miR-99b-3p | 3.130 | 1.659 | 0.250 | Not DEG |
| rno-miR-19b-1-5p | 0.526 | 1.630 | 0.616 | Not DEG |
| rno-miR-463-3p | 1.364 | 1.577 | 0.424 | Not DEG |
| rno-miR-540-5p | 0.469 | 1.518 | 0.623 | Not DEG |
| rno-miR-211-5p | 6.437 | 1.517 | 0.183 | Not DEG |
| rno-miR-98-5p | 24.956 | 1.482 | 0.154 | Not DEG |
baseMean: the average levels of miRNA expression; log2 (fold change): the 2-logarithm value of the expression fold change of certain miRNA; DEG: differentially expressed genes; NA: not analyzed
Supplementary Table 6.
A collection of top twenty downregulated miRNAs detected by next-generation sequencing in prostate tissues (blank control group vs normal saline group)
| miRNA | baseMean | log2 (fold change) | P | type |
|---|---|---|---|---|
| rno-miR-206-3p | 12.472 | 6.663 | NA | Not DEG |
| rno-miR-124-3p | 20.447 | 4.075 | 0.002 | Down |
| rno-miR-3559-5p | 381.091 | 3.973 | NA | Not DEG |
| rno-miR-673-5p | 0.577 | 3.970 | 0.203 | Not DEG |
| rno-miR-17-2-3p | 0.551 | 3.922 | 0.264 | Not DEG |
| rno-miR-3064-5p | 0.520 | 3.873 | 0.229 | Not DEG |
| rno-miR-493-5p | 0.357 | 3.508 | 0.348 | Not DEG |
| rno-miR-3583-3p | 0.326 | 3.448 | 0.388 | Not DEG |
| rno-miR-673-3p | 0.817 | 3.402 | 0.188 | Not DEG |
| rno-miR-101b-3p | 9730.208 | 3.364 | NA | Not DEG |
| rno-miR-1-3p | 156.879 | 3.289 | NA | Not DEG |
| rno-miR-6331 | 0.276 | 3.270 | 0.413 | Not DEG |
| rno-miR-326-5p | 0.276 | 3.270 | 0.413 | Not DEG |
| rno-miR-298-5p | 0.276 | 3.270 | 0.413 | Not DEG |
| rno-miR-380-3p | 0.276 | 3.270 | 0.413 | Not DEG |
| rno-miR-1298 | 0.245 | 3.191 | 0.425 | Not DEG |
| rno-miR-136-3p | 4.326 | 3.189 | 0.070 | Not DEG |
| rno-miR-532-5p | 466.241 | 3.122 | NA | Not DEG |
| rno-miR-598-3p | 4.106 | 3.057 | 0.109 | Not DEG |
| rno-miR-129-2-3p | 0.163 | 2.853 | 0.477 | Not DEG |
baseMean: the average levels of miRNA expression; log2 (fold change): the 2-logarithm value of the expression fold change of certain miRNA; DEG: differentially expressed gene; NA: not analyzed
Validation of differentially expressed miRNAs revealed by NGS
To better evaluate the accuracy of NGS data, we have established a new batch of CNP model rats and corresponding control groups. Validation of altered miRNA expression in CNP tissues profiled by NGS was performed by qRT-PCR in BC, NS, and CAR groups. We randomly picked five upregulated and five downregulated miRNAs among the differentially expressed miRNAs to conduct the analysis, and each expression abundance was presented using the normalized fold change of CAR group versus BC and NS groups. As a result, the altered expression tendency of all the ten deregulated miRNAs was consistent with the data obtained from NGS technology. Besides, overwhelming majority of the tested miRNAs showed significant differences between CAR versus BC and CAR versus NS. Among all the compared miRNAs, significant differences were only not found in miR-130a-3p and miR-26a-3p between CAR and NS, which together indicated that the expression levels of miRNAs detected by NGS were reliable (Figure 3).
Figure 3.
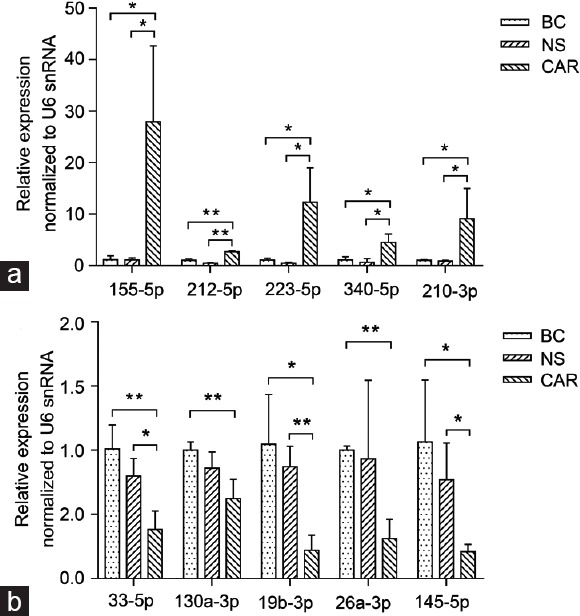
Relative expression of (a) five upregulated and (b) five downregulated miRNAs in BC, NS, and CAR groups at day 7 after model establishment (n=3 for each group). Data are presented as mean ± s.e.m. *P < 0.05, **P < 0.01 BC: blank control group; NS: normal saline group; CAR: carrageenan group; s.e.m: standard error of the mean.
GO enrichment analysis of differentially expressed genes in the CNP prostate tissues
The essential biological functions of the putative target genes were classified via the GO analysis. To better understand the biological behaviors of the known deregulated miRNAs, potential target genes and biological functions targeted by these miRNAs were predicted. The top ten most enriched GO terms for biological processes (BPs), cellular components (CCs), and molecular functions (MFs) are presented in Supplementary Table 7, 8 and Supplementary Figure 4 (3.6MB, tif) . Of these genes, the putative target genes of deregulated miRNAs appeared to be mostly enriched for CC such as cell, cell part, intracellular, intracellular part, and organelle, when comparing the CAR group with either the BC or NS group. Meanwhile, the most enriched MFs were binding, protein binding, ion binding, organic cyclic compound binding, and heterocyclic compound binding, and the most enriched BPs were cellular process, single-organism process, metabolic process, organic substance metabolic process, and cellular metabolic process for the CAR versus BC or CAR versus NS comparison. Correspondingly, the putative target genes of deregulated miRNAs between CAR and NS appeared to mostly overlap with the CAR versus BC comparison.
Supplementary Table 7.
Top ten most enriched gene ontology terms for predicted targets of differentially expressed miRNAs (carrageenan group vs blank control group)
| ID | Description | GeneRatio | P | Type |
|---|---|---|---|---|
| GO:0005623 | Cell | 3618/3978 | 4.171E-114 | CC |
| GO:0044464 | Cell part | 3609/3978 | 2.588E-115 | CC |
| GO:0009987 | Cellular process | 3488/3838 | 2.116E-100 | BP |
| GO:0005622 | Intracellular | 3248/3978 | 3.592E-152 | CC |
| GO:0044699 | Single-organism process | 3222/3838 | 7.145E-92 | BP |
| GO:0044424 | Intracellular part | 3138/3978 | 6.305E-141 | CC |
| GO:0043226 | Organelle | 2951/3978 | 1.539E-105 | CC |
| GO:0005488 | Binding | 2895/3655 | 9.936E-155 | MF |
| GO:0044763 | Single-organism cellular process | 2911/3838 | 9.523E-66 | BP |
| GO:0043227 | Membrane-bounded organelle | 2780/3978 | 2.099E-115 | CC |
ID: the label information in GO database; Description: the descriptive information of GO function; GeneRatio: the proportion of GO term-related genes of total differential genes; BP: biological process; CC: cellular component; MF: molecular function; GO: gene ontology
Supplementary Table 8.
Top ten most enriched gene ontology terms for predicted targets of differentially expressed miRNAs (carrageenan group vs normal saline group)
| ID | Description | GeneRatio | P | Type |
|---|---|---|---|---|
| GO:0005623 | Cell | 3273/3601 | 1.795E-100 | CC |
| GO:0044464 | Cell part | 3265/3601 | 1.290E-101 | CC |
| GO:0009987 | Cellular process | 3137/3467 | 9.759E-83 | BP |
| GO:0005622 | Intracellular | 2945/3601 | 2.697E-137 | CC |
| GO:0044699 | Single-organism process | 2886/3467 | 1.820E-72 | BP |
| GO:0044424 | Intracellular part | 2838/3601 | 6.137E-124 | CC |
| GO:0043226 | Organelle | 2696/3601 | 1.333E-102 | CC |
| GO:0005488 | Binding | 2642/3324 | 1.329E-142 | MF |
| GO:0044763 | Single-organism cellular process | 2606/3467 | 9.910E-52 | BP |
| GO:0043227 | Membrane-bounded organelle | 2542/3601 | 1.142E-111 | CC |
ID: the label information in GO database; Description: the descriptive information of GO function; GeneRatio: the proportion of GO term-related genes of total differential genes; BP: biological process; CC: cellular component; MF: molecular function; GO: gene ontology
KEGG pathway analysis of differentially expressed genes in the CNP prostate tissues
After GO analysis, the putative target genes of the differentially expressed miRNAs in the prostate specimens were examined by KEGG for pathway enrichment analysis. The top ten enriched KEGG pathways are presented in Figure 4. Based on the proportion of differentially expressed genes in all the annotated genes, Q-values (adjusted P values), and number of genes enriched in specific pathways, we have annotated the most enriched pathways. When comparing CAR with BC, these pathways were tightly linked to cancer, endocytosis, proteoglycans in cancer, axon guidance, and forkhead box O (FoxO). When comparing CAR with NS, these pathways were tightly linked to cancer, mitogen-activated protein kinase (MAPK), proteoglycans in cancer, cyclic adenosine monophosphate (cAMP), and calcium signaling pathways. Furthermore, to confirm that the aforementioned deregulated miRNAs could contribute to affecting the enriched KEGG pathways, we have selected two essential genes cAMP and mammalian target of rapamycin (mTOR) for mRNA expression validation from “cAMP signaling pathway” and “mTOR signaling pathway.” As a result, cAMP showed significant increased expression in CAR group, while mTOR presented significant decreased expression in CAR group, compared with BC and NS groups (P < 0.01; Supplementary Figure 5 (414.1KB, tif) ). These results indicated that miRNA expression changes in fact caused altered gene expression in enriched pathway, which may further initiate the activation or depression of the following signaling transduction involved in inflammation and immune response.
Figure 4.
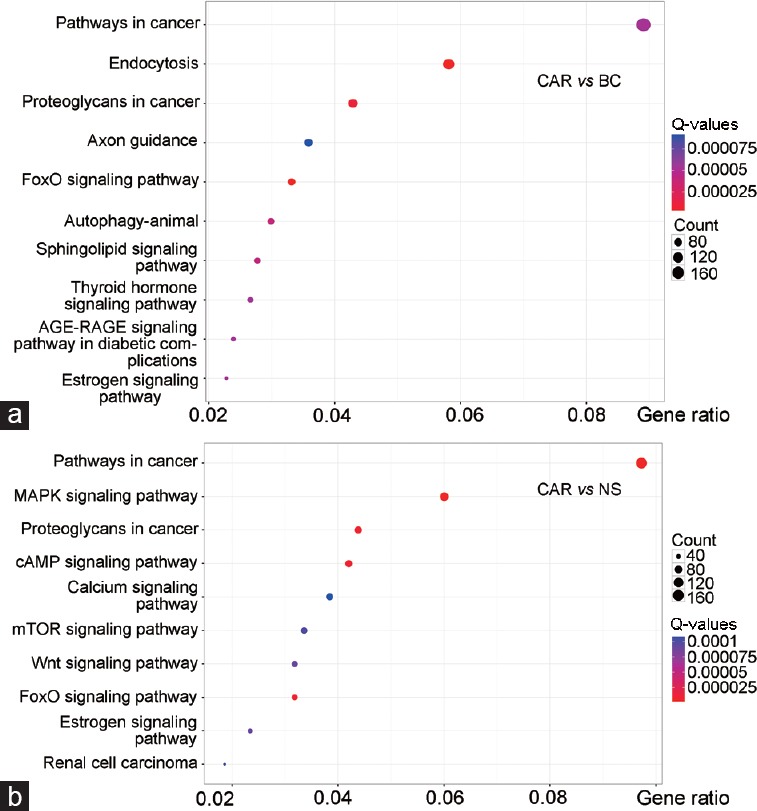
KEGG enrichment dot plot for target genes of differentially expressed miRNAs (CAR vs BC and CAR vs NS). The top ten enriched KEGG pathways are presented. The X-axis represents enrichment factor, indicating the proportion of differentially expressed genes in all annotated genes on certain signaling pathway. The Y-axis represents log10 (Q-values). Enriched pathways with higher X and Y values possessed better statistical significances. (a) CAR versus BC; (b) CAR versus NS. BC: blank control group; NS: normal saline group; CAR: carrageenan group; KEGG: Kyoto Encyclopedia of Genes and Genomes; FoxO: forkhead box O; AGE-RAGE: advanced glycation end products-receptor for advanced glycation end products; MAPK: mitogen-activated protein kinase; cAMP: cyclic adenosine monophosphate; mTOR: mammalian target of rapamycin; Wnt: wingless/int 1; Q-values: adjusted P values.
DISCUSSION
CNP is a complex and frustrating syndrome caused by unknown pathological mechanisms with a lack of effective therapeutics. Massive efforts have been made to uncover the pivotal factors that promote the occurrence and development of CNP. Although only one-third have inflammation on biopsy,29 and how chronic pelvic pain develops and whether it could be mechanistically linked with inflammation were still controversial, the crucial role of inflammation in CNP and symptoms such as chronic pelvic pain should not be unnoticed, as unresolved chronic inflammation may potentiate tissue injury resulting in central sensitization and eventually cause chronic pelvic pain.10,30,31 Recently, researches in biomedical science studies have indicated that inflammation and immune responses coexist in various kinds of diseases.32 Correspondingly, immune mechanisms have been reported to be involved in CNP etiology, pathogenesis, and symptom development.10 However, full understanding of the exact molecular mechanisms, especially the immune regulatory networks involved in CNP, remains largely absent. Notably, an increasing number of studies have shown that microRNAs, a class of evolutionally conserved and endogenously expressed RNAs, could be considered as critical regulators of gene expression at posttranscriptional levels.33 MiRNAs are involved in the pathogenesis of a broad spectrum of human disorders, including metabolic and inflammatory diseases.34 Accordingly, as CNP has shown complex symptom features such as prostate inflammation and immune disorders, it is of great importance to generate the differentially expressed miRNA profiles in prostatitis tissues to further understand the miRNA regulatory networks in CNP initiation and progression, especially to clearly understand the possible role of the immune system in CNP.
Recently, a newly developed NGS technology has been widely used for miRNA quantitative studies, due to the higher test specificity and sensitivity to detect extremely low-abundance miRNAs that may not be detected by the traditional quantitative assays. Based on NGS, bioinformatics analyses of deregulated miRNA expression signatures have further enriched several significant GO terms and signaling pathways, which are considered to be related to inflammatory diseases such as osteoarthritis35 and rheumatoid arthritis (RA).36 However, no systematic studies have been conducted on miRNAs profiling in CNP tissues by NGS.
In the present study, we used NGS technology to identify miRNAs prepared from prostate tissues from a CNP animal model. We quantified a total of 20 048 744, 15 070 023, and 9 183 863 sequencing read counts with typical 18–36 nts length for the BC, NS, and CAR groups, respectively, following alignment to the rat genome sequence dataset, which represented 15 526 924, 12 752 646, and 7 918 591 read counts aligned to miRBase for each group (n = 3), respectively. Among these alignments, 1224, 1039, and 1029 genome-wide expressed known miRNAs accompanied by 257, 204, and 190 novel miRNAs were in BC, NS, and CAR groups, respectively. As the histological origins of paired samples undergoing NGS were identical, most of the top twenty highly expressed miRNAs in the three groups showed similar expression pattern, while rno-miR-146a-5p was found to be differentially expressed between BC and CAR groups (P < 0.001) and also between NS and CAR groups (P < 0.001). Interestingly, Figure 3 showed that CAR treatment group had a far higher abundance of tRNAs identified (7.54%) than BC (2.60%) or NS (1.48%) groups. tRNAs can be processed to distinct small RNA species commonly referred to as tRNA-derived fragments (tRFs or tDRs). Based on the origin from 5' or 3' end, tRFs were divided into tRF-5 and tRF-3.37 It has been revealed that a particular tRF-3 (named CU1276 or tRF-3027) was downregulated in human malignant B-cells and lymphoma biopsies compared with naive, germinal, and memory B-cells. It interacted with argonaute (AGO) proteins, and its overexpression could repress the endogenous abundance of the tumor suppressor replication protein A1 (RPA1) in a miRNA-like manner.38 These results indicated that CAR treatment might cause the deregulated expression of several posttranscriptional modulators including not only miRNAs but also some tRNAs.
Furthermore, based on a “volcano plot” that simultaneously defined small P values and high fold changes, we focused on exploring miRNAs that showed significantly different expression patterns. The comparative analysis identified 84 and 70 dysregulated miRNAs between the CAR group and BC group, and between CAR group and NS group, respectively. Notably, of the top twenty differentially upregulated or downregulated miRNAs in CAR versus BC and CAR versus NS comparisons, 14 miRNAs were overlapped either in upregulated or downregulated group, indicating the consistent tendencies after the injection of carrageenan.
To explore the concrete regulatory roles of the overlapped and dysregulated miRNAs in CNP, we firstly compared the expression patterns between our results and those of previous publications. As mentioned before, it has been reported that miR-146a rs2910164 C allele could compromise miR-146a expression and thereby increase the expression of nitric oxide synthase 1 (NOS1) for decreased risk of ED.15 In consideration of that ED was substantially increased in patients with CP, these findings were consistent with our data that miR-146a-5p/3p was upregulated in carrageenan-induced prostatitis. Besides, miR-141 was addressed to be downregulated in CP patients,16 again verifying our similar findings in carrageenan-induced prostatitis model (Supplementary Table 4). Later on, we have also attempted to link our results to those reports regarding other inflammatory diseases. For example, miR-146b-5p has been reported to be highly expressed in inflamed temporal artery biopsies from patients with giant cell arteritis;39 it was also significantly elevated in CNP tissues in our NGS data. miR-511-3p, which is associated with macrophage-mediated pro-inflammatory cytokine production and contributed to inflammation,40 was significantly upregulated in our investigations. A similar result was obtained for upregulated miR-155-5p in endomyocardial patients with inflammatory cardiomyopathy.41 Notably, miR-155-5p was regarded as a critical component in an inflammation-induced regulatory loop that controlled T-cell function and peripheral immune tolerance.42 Thus, upregulated miR-155-5p may promote the progression of CNP inflammation through the enhancement of peripheral immune tolerance.
Among the representative downregulated miRNAs in the CAR group, miR-139-5p was also significantly decreased in clinically advanced primary biliary cholangitis.43 It was also found that miR-200b was significantly downregulated in early pulmonary inflammation and fibrosis development, which was the same as its expression pattern reported in CNP.44 Specifically, miR-150-5p, which was decreased in enterovirus-induced inflammatory injury in central nervous system,45 was significantly downregulated in the CAR group. Given that extracellular vesicles released by CD4+ T helper (Th) lymphocytes such as T regulatory (Treg) cells contained enriched miR-150-5p levels to inhibit CD4+ T-cell proliferation,46 the decreased exocrine miR-150-5p levels in Treg cells could be one of the possible mechanisms leading to inflammation. More importantly, we have identified several new miRNAs with altered expression, including miR-1956-3p, miR-6215, miR-883-5p, and miR-1247-5p, whose roles in inflammation were largely unknown before.
Intriguingly, there were certain dysregulated miRNAs identified in the current work that showed inconsistent expression patterns with published data. For example, miR-19b-3p was upregulated in plasma from knee osteoarthritis patients and was positively correlated with disease severity.35 However, the expression of miR-19b-3p in CNP was decreased. The probable explanation was that miRNAs might exert distinct expression levels in plasma or local tissue; additionally, the complex pathological mechanisms and regulatory networks of miRNAs may not be uniform in different kinds of inflammatory diseases.
Following the differential expression analysis, we performed GO and KEGG bioinformatics analyses. The significantly enriched GO terms revealed the participation of target genes in several biological processes such as regulation of immune system process, immune response, immune effector process, inflammatory response, innate immune response, and activation of immune response, indicating the regulatory effects of miRNAs in inflammation progression and immune responses. KEGG pathway analysis enriched several immune-related pathways, such as “MAPK signaling pathway,” “cAMP signaling pathway,” “endocytosis pathway,” “mTOR signaling pathway,” and “FoxO signaling pathway.” Generally, MAPK signaling pathway is thought to be involved in innate immune Toll-like receptor (TLR) expression47 and is important in the T-cell antigen receptor (TCR) signaling negative feedback loop.48 The cAMP signaling network is necessary for inducing the catalytic activity of protein kinase A (PKA) in splenocytes, lymphocytes, and thymocytes, which is important for regulating innate as well as adaptive immune sensitivities.49 cAMP was the first identified intracellular second messenger of extracellular ligand action. Within the immune system, elevated intracellular cAMP levels generally reduce pro-inflammatory mediators and increase anti-inflammatory factor production,50 while among T-cells, Treg produce and respond to prostaglandin E2 (PGE2), which is modulated by COX-2 and acts as an autocrine factor to increase intracellular cAMP. It has been reported that nonsteroidal anti-inflammatory drugs (NSAIDs) and COX-2 inhibitors could block PGE2 synthesis, eventually downregulating intracellular cAMP levels in T-cells, suggesting the crucial role of cAMP in maintaining the immune balance.51 We found in our study that cAMP mRNA expression and pro-inflammatory miR-155-5p were both upregulated in CNP after CAR treatment, this was consistent with a previous report that cAMP and miR-155-5p may modulate inflammatory process.52 Similarly, the enriched endocytosis pathway also has been reported to participate in enhanced major histocompatibility complex II (MHC II) presentation mediated by CD4+ T-cells.53 mTOR complex 1 (mTORC1)-induced glycolytic reprograming can activate human monocytes and macrophages and subsequently promote pro-inflammatory cytokine production.54 mTOR is a serine/threonine kinase that regulates cellular growth, proliferation, inflammation, and immune in response to various environmental stimuli. Although the concrete roles of mTOR in inflammation and immune were largely controversial, a central role for mTOR in innate immunity has been recently defined by its ability to restrain pro-inflammatory factors. It has been reported that inhibition of mTOR by rapamycin could augment inflammation and pulmonary injury through NF-κB activity enhancement.55 Similarly, in myeloid immune cells, mTOR inhibition was demonstrated to effectively block the anti-inflammatory effects of glucocorticoids.56 In our current investigation, as expected, mTOR downregulation was observed with upregulated miR-155-5p in CAR-induced CNP, which was supported by the fact that miR-155-5p could inhibit mTOR expression,57 together indicating that mTOR and miR-155-5p may coordinately modulate CNP through complex network involved in immune cells. Meanwhile, the FoxO pathway is considered to promote immune activity by negatively regulating the expression of immunosuppressive proteins including programmed death receptor 1 ligand (PD-L1).58 Thus, our results suggest that immune factors are probably involved in CNP regulation networks facilitated by miRNAs.
CONCLUSION
We have obtained the first data for the comprehensive characterization of miRNAs in CNP prostate tissues with NGS technology and identified several differentially expressed miRNAs that are potentially associated with prostatic inflammation progression. We have also enriched several immune-associated pathways, indicating the possible immunological regulatory functions in CNP progression. Our data provide an important platform for future investigations aiming to characterize the concrete roles of specific miRNAs and immune factors in CNP initiation, progression, and the development of effective theragnosis.
AUTHOR CONTRIBUTIONS
CZL and LZ designed this experiment. LZ, YL, XGC, YZ, JC, LGZ, and HXD performed the research and analyzed the data. YL and ZYH wrote the manuscript, and then LZ, SF, and CZL revised the manuscript. All authors read and approved the final manuscript.
COMPETING INTERESTS
All authors declare no competing interests.
HE histological staining and IHC staining of COX-2 in prostate tissues. Histological staining pictures (a–l) of prostate at the first day (a and d for BC; b and e for NS; c and f for CAR) and the 7th day (g and j for BC; h and k for NS; i and l for CAR) among the three groups are presented as above. Numerous infiltrated inflammatory cells have been found in prostatic stroma at the 7th day after carrageenan injection. Meanwhile, IHC images (m–x) of COX-2 at the first day (m and p for BC; n and q for NS; o and r for CAR) and the 7th day (s and v for BC; t and w for NS; u and x for CAR) are presented, the higher magnification (×400) and local representative visual area are indicated by white arrows as well as magnification in box. The expression of COX-2 was significantly increased in prostatic glandular epithelium and infiltrated cells in stroma. The scale bars are consistently 200 µm. COX-2: cyclooxygenase-2; BC: blank control group; NS: normal saline group; CAR: carrageenan group.
COX-2 expression in BC, NS, and CAR groups at day 1 and day 7. The mean densities of prostate tissues from three cases were presented at the top graph and summarized in the bottom table. Mean ± s.e.m, n = 3, #P > 0.05, **P < 0.01, ***P < 0.001, compared with the control of each group. COX-2: cyclooxygenase-2; BC: blank control group; NS: normal saline group; CAR: carrageenan group; s.e.m: standard error of the mean.
Increased IFN-γ in rats with carrageenan-induced CNP. Circulating IFN-γ levels in serum samples of BC, NS, and CAR groups on day 7 after model establishment. Mean ± s.e.m, n = 3, ***P < 0.001, compared with BC and NS groups. IFN-γ: interferon-γ; BC: blank control group; NS: normal saline group; CAR: carrageenan group; s.e.m: standard error of the mean; CNP: chronic nonbacterial prostatitis.
GO enrichment bar plot of target genes of differentially expressed miRNA. The top ten enriched GO functions (based on P values) in separated categories of BP, CC, and MF. The number at the end of each column represents the number of differentially expressed genes in the corresponding enriched GO function, which could reflect the gene distribution on certain GO functions. (a) CAR versus BC; (b) CAR versus NS. BC: blank control group; NS: normal saline group; CAR: carrageenan group; GO: gene Ontology; BP: biological process; CC: cellular component; MF: molecular function.
Relative expression of cAMP and mTOR in BC, NS, and CAR groups at day 7. Mean ± s.e.m., n = 3, **P < 0.01, compared with the control of each group. BC: blank control group; NS: normal saline group; CAR: carrageenan group; cAMP: cyclic adenosine monophosphate; mTOR: mammalian target of rapamycin; s.e.m: standard error of the mean.
ACKNOWLEDGMENTS
We are grateful to Dr. Yu Yin at the First Affiliated Hospital of Anhui Medical University (AHMU) for her technical assistance in histological research. This work was supported by the National Natural Science Foundation of China (Grant No. 81630019, 31430028, 81401518, and 81470986), Anhui Provincial Institutes for Translational Medicine (Grant No. 2017ZHYX02), Cultivation Project of Young Top-Notch Talent Support from Anhui Medical University (AHMU), and Funding for Distinguished Young Scientists of the First Affiliated Hospital of AHMU.
Supplementary Information is linked to the online version of the paper on the Asian Journal of Andrology website.
REFERENCES
- 1.Eltzschig HK, Carmeliet P. Hypoxia and inflammation. N Engl J Med. 2011;364:656–65. doi: 10.1056/NEJMra0910283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Netea MG, Balkwill F, Chonchol M, Cominelli F, Donath MY, et al. A guiding map for inflammation. Nat Immunol. 2017;18:826–31. doi: 10.1038/ni.3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dinarello CA. Historical insights into cytokines. Eur J Immunol. 2007;37(Suppl 1):S34–45. doi: 10.1002/eji.200737772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Artis D, Spits H. The biology of innate lymphoid cells. Nature. 2015;517:293–301. doi: 10.1038/nature14189. [DOI] [PubMed] [Google Scholar]
- 5.Firestein GS, McInnes IB. Immunopathogenesis of rheumatoid arthritis. Immunity. 2017;46:183–96. doi: 10.1016/j.immuni.2017.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Noda S, Suarez-Farinas M, Ungar B, Kim SJ, de Guzman Strong C, et al. The Asian atopic dermatitis phenotype combines features of atopic dermatitis and psoriasis with increased TH17 polarization. J Allergy Clin Immunol. 2015;136:1254–64. doi: 10.1016/j.jaci.2015.08.015. [DOI] [PubMed] [Google Scholar]
- 7.Dietze-Schwonberg K, Lorenz B, Kostka SL, Schumak B, Gessner A, et al. Insufficient generation of TH17 cells in IL-23p19-deficient BALB/c mice protects against progressive cutaneous leishmaniasis. Exp Dermatol. 2018;27:101–3. doi: 10.1111/exd.13455. [DOI] [PubMed] [Google Scholar]
- 8.Lai H, Gereau RW, Luo Y, O'Donnell M, Rudick CN, et al. Animal models of urologic chronic pelvic pain syndromes: findings from the multidisciplinary approach to the study of chronic pelvic pain research network. Urology. 2015;85:1454–65. doi: 10.1016/j.urology.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holt JD, Garrett WA, McCurry TK, Teichman JM. Common questions about chronic prostatitis. Am Fam Physician. 2016;93:290–6. [PubMed] [Google Scholar]
- 10.Breser ML, Salazar FC, Rivero VE, Motrich RD. Immunological mechanisms underlying chronic pelvic pain and prostate inflammation in chronic pelvic pain syndrome. Front Immunol. 2017;8:898. doi: 10.3389/fimmu.2017.00898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murphy SF, Schaeffer AJ, Thumbikat P. Immune mediators of chronic pelvic pain syndrome. Nat Rev Urol. 2014;11:259–69. doi: 10.1038/nrurol.2014.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Motrich RD, Maccioni M, Molina R, Tissera A, Olmedo J, et al. Presence of INFgamma-secreting lymphocytes specific to prostate antigens in a group of chronic prostatitis patients. Clin Immunol. 2005;116:149–57. doi: 10.1016/j.clim.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 13.Pan W, Zhu S, Dai D, Liu Z, Li D, et al. MiR-125a targets effector programs to stabilize Treg-mediated immune homeostasis. Nat Commun. 2015;6:7096. doi: 10.1038/ncomms8096. [DOI] [PubMed] [Google Scholar]
- 14.Mann M, Mehta A, Zhao JL, Lee K, Marinov GK, et al. An NF-κB-microRNA regulatory network tunes macrophage inflammatory responses. Nat Commun. 2017;8:851. doi: 10.1038/s41467-017-00972-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ding J, Tang Y, Tang Z, Zhang X, Wang G. A variant in the precursor of microRNA-146a is responsible for development of erectile dysfunction in patients with chronic prostatitis via targeting NOS1. Med Sci Monit. 2017;23:929–37. doi: 10.12659/MSM.898406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang LL, Huang YH, Yan CY, Wei XD, Hou JQ, et al. N-acetylcysteine ameliorates prostatitis via miR-141 regulating Keap1/Nrf2 signaling. Inflammation. 2016;39:938–47. doi: 10.1007/s10753-016-0327-1. [DOI] [PubMed] [Google Scholar]
- 17.Chen Y, Chen S, Zhang J, Wang Y, Jia Z, et al. Expression profile of microRNAs in expressed prostatic secretion of healthy men and patients with IIIA chronic prostatitis/chronic pelvic pain syndrome. Oncotarget. 2018;9:12186–200. doi: 10.18632/oncotarget.24069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeng F, Chen H, Yang J, Wang L, Cui Y, et al. Development and validation of an animal model of prostate inflammation-induced chronic pelvic pain: evaluating from inflammation of the prostate to pain behavioral modifications. PLoS One. 2014;9:e96824. doi: 10.1371/journal.pone.0096824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Calio A, Eble JN, Hes O, Martignoni G, Harari SE, et al. Distinct clinicopathological features in metanephric adenoma harboring BRAF mutation. Oncotarget. 2017;8:54096–105. doi: 10.18632/oncotarget.11117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tong XH, Xu B, Zhang YW, Liu YS, Ma CH. Research resources: comparative microRNA profiles in human corona radiata cells and cumulus oophorus cells detected by next-generation small RNA sequencing. PLoS One. 2014;9:e106706. doi: 10.1371/journal.pone.0106706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nawrocki EP, Burge SW, Bateman A, Daub J, Eberhardt RY, et al. Rfam 12.0: updates to the RNA families database. Nucleic Acids Res. 2015;43:D130–7. doi: 10.1093/nar/gku1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Friedlander MR, Chen W, Adamidi C, Maaskola J, Einspanier R, et al. Discovering microRNAs from deep sequencing data using miRDeep. Nat Biotechnol. 2008;26:407–15. doi: 10.1038/nbt1394. [DOI] [PubMed] [Google Scholar]
- 24.Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–17. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA. org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–53. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–4. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Motrich RD, van Etten E, Baeke F, Riera CM, Mathieu C, et al. Crucial role of interferon-gamma in experimental autoimmune prostatitis. J Urol. 2010;183:1213–20. doi: 10.1016/j.juro.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 28.Perez-Alvarado C, Gomez C, Reyes M, Garcia M, Perez E, et al. Anti-inflammatory effect of dialyzable leukocyte extract in autoimmune prostatitis: evaluation in animal model. Biomed Res Int. 2017;2017:1832853. doi: 10.1155/2017/1832853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.True LD, Berger RE, Rothman I, Ross SO, Krieger JN. Prostate histopathology and the chronic prostatitis/chronic pelvic pain syndrome: a prospective biopsy study. J Urol. 1999;162:2014–8. doi: 10.1016/S0022-5347(05)68090-1. [DOI] [PubMed] [Google Scholar]
- 30.Watanabe T, Inoue M, Sasaki K, Araki M, Uehara S, et al. Nerve growth factor level in the prostatic fluid of patients with chronic prostatitis/chronic pelvic pain syndrome is correlated with symptom severity and response to treatment. BJU Int. 2011;108:248–51. doi: 10.1111/j.1464-410X.2010.09716.x. [DOI] [PubMed] [Google Scholar]
- 31.Nickel JC, Roehrborn CG, O'Leary MP, Bostwick DG, Somerville MC, et al. Examination of the relationship between symptoms of prostatitis and histological inflammation: baseline data from the REDUCE chemoprevention trial. J Urol. 2007;178:896–900. doi: 10.1016/j.juro.2007.05.041. [DOI] [PubMed] [Google Scholar]
- 32.Marques RE, Marques PE, Guabiraba R, Teixeira MM. Exploring the homeostatic and sensory roles of the immune system. Front Immunol. 2016;7:125. doi: 10.3389/fimmu.2016.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gommans WM, Berezikov E. Controlling miRNA regulation in disease. Methods Mol Biol. 2012;822:1–18. doi: 10.1007/978-1-61779-427-8_1. [DOI] [PubMed] [Google Scholar]
- 34.Zhong H, Ma M, Liang T, Guo L. Role of microRNAs in obesity-induced metabolic disorder and immune response. J Immunol Res. 2018;2018:2835761. doi: 10.1155/2018/2835761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kong R, Gao J, Si Y, Zhao D. Combination of circulating miR-19b-3p, miR-122-5p and miR-486-5p expressions correlates with risk and disease severity of knee osteoarthritis. Am J Transl Res. 2017;9:2852–64. [PMC free article] [PubMed] [Google Scholar]
- 36.Rajasekhar M, Olsson AM, Steel KJ, Georgouli M, Ranasinghe U, et al. MicroRNA-155 contributes to enhanced resistance to apoptosis in monocytes from patients with rheumatoid arthritis. J Autoimmun. 2017;79:53–62. doi: 10.1016/j.jaut.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs) Genes Dev. 2009;23:2639–49. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maute RL, Schneider C, Sumazin P, Holmes A, Califano A, et al. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc Natl Acad Sci USA. 2013;110:1404–9. doi: 10.1073/pnas.1206761110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Croci S, Zerbini A, Boiardi L, Muratore F, Bisagni A, et al. MicroRNA markers of inflammation and remodelling in temporal arteries from patients with giant cell arteritis. Ann Rheum Dis. 2016;75:1527–33. doi: 10.1136/annrheumdis-2015-207846. [DOI] [PubMed] [Google Scholar]
- 40.Heinsbroek SE, Squadrito ML, Schilderink R, Hilbers FW, Verseijden C, et al. miR-511-3p, embedded in the macrophage mannose receptor gene, contributes to intestinal inflammation. Mucosal Immunol. 2016;9:960–73. doi: 10.1038/mi.2015.113. [DOI] [PubMed] [Google Scholar]
- 41.Besler C, Urban D, Watzka S, Lang D, Rommel KP, et al. Endomyocardial miR-133a levels correlate with myocardial inflammation, improved left ventricular function, and clinical outcome in patients with inflammatory cardiomyopathy. Eur J Heart Fail. 2016;18:1442–51. doi: 10.1002/ejhf.579. [DOI] [PubMed] [Google Scholar]
- 42.Yee D, Shah KM, Coles MC, Sharp TV, Lagos D. MicroRNA-155 induction via TNF-α and IFN-γ suppresses expression of programmed death ligand-1 (PD-L1) in human primary cells. J Biol Chem. 2017;292:20683–93. doi: 10.1074/jbc.M117.809053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Katsumi T, Ninomiya M, Nishina T, Mizuno K, Tomita K, et al. miR-139-5p is associated with inflammatory regulation through c-FOS suppression, and contributes to the progression of primary biliary cholangitis. Lab Invest. 2016;96:1165–77. doi: 10.1038/labinvest.2016.95. [DOI] [PubMed] [Google Scholar]
- 44.Cao Y, Liu Y, Ping F, Yi L, Zeng Z, et al. miR-200b/c attenuates lipopolysaccharide-induced early pulmonary fibrosis by targeting ZEB1/2 via p38 MAPK and TGF-β/smad3 signaling pathways. Lab Invest. 2018;98:339–59. doi: 10.1038/labinvest.2017.123. [DOI] [PubMed] [Google Scholar]
- 45.Yang X, Xie J, Jia L, Liu N, Liang Y, et al. Analysis of miRNAs involved in mouse brain damage upon enterovirus 71 infection. Front Cell Infect Microbiol. 2017;7:133. doi: 10.3389/fcimb.2017.00133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Torri A, Carpi D, Bulgheroni E, Crosti MC, Moro M, et al. Extracellular microRNA signature of human helper T cell subsets in health and autoimmunity. J Biol Chem. 2017;292:2903–15. doi: 10.1074/jbc.M116.769893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sindhu S, Akhter N, Kochumon S, Thomas R, Wilson A, et al. Increased expression of the innate immune receptor TOL10 in obesity and type-2 diabetes: association with ROS-mediated oxidative stress. Cell Physiol Biochem. 2018;45:572–90. doi: 10.1159/000487034. [DOI] [PubMed] [Google Scholar]
- 48.Giardino Torchia ML, Dutta D, Mittelstadt PR, Guha J, Gaida MM, et al. Intensity and duration of TCR signaling is limited by p38 phosphorylation of ZAP-70T293 and destabilization of the signalosome. Proc Natl Acad Sci U S A. 2018;115:2174–9. doi: 10.1073/pnas.1713301115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moen LV, Sener Z, Volchenkov R, Svarstad AC, Eriksen AM, et al. Ablation of the Cβ2 subunit of PKA in immune cells leads to increased susceptibility to systemic inflammation in mice. Eur J Immunol. 2017;47:1880–9. doi: 10.1002/eji.201646809. [DOI] [PubMed] [Google Scholar]
- 50.Raker VK, Becker C, Steinbrink K. The cAMP pathway as therapeutic target in autoimmune and inflammatory diseases. Front Immunol. 2016;7:123. doi: 10.3389/fimmu.2016.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wehbi VL, Tasken K. Molecular mechanisms for cAMP-mediated immunoregulation in T cells-role of anchored protein kinase a signaling units. Front Immunol. 2016;7:222. doi: 10.3389/fimmu.2016.00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fassi Fehri L, Koch M, Belogolova E, Khalil H, Bolz C, et al. Helicobacter pylori induces miR-155 in T cells in a cAMP-Foxp3-dependent manner. PLoS One. 2010;5:e9500. doi: 10.1371/journal.pone.0009500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dixit S, Sahu R, Verma R, Duncan S, Giambartolomei GH, et al. Caveolin-mediated endocytosis of the Chlamydia M278 outer membrane peptide encapsulated in poly(lactic acid)-Poly(ethylene glycol) nanoparticles by mouse primary dendritic cells enhances specific immune effectors mediated by MHC class II and CD4+ T cells. Biomaterials. 2018;159:130–45. doi: 10.1016/j.biomaterials.2017.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yoon BR, Oh YJ, Kang SW, Lee EB, Lee WW. Role of SLC7A5 in metabolic reprogramming of human monocyte/macrophage immune responses. Front Immunol. 2018;9:53. doi: 10.3389/fimmu.2018.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yoshida T, Mett I, Bhunia AK, Bowman J, Perez M, et al. Rtp801, a suppressor of mTOR signaling, is an essential mediator of cigarette smoke-induced pulmonary injury and emphysema. Nat Med. 2010;16:767–73. doi: 10.1038/nm.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weichhart T, Haidinger M, Katholnig K, Kopecky C, Poglitsch M, et al. Inhibition of mTOR blocks the anti-inflammatory effects of glucocorticoids in myeloid immune cells. Blood. 2011;117:4273–83. doi: 10.1182/blood-2010-09-310888. [DOI] [PubMed] [Google Scholar]
- 57.Wang F, Shan S, Huo Y, Xie Z, Fang Y, et al. MiR-155-5p inhibits PDK1 and promotes autophagy via the mTOR pathway in cervical cancer. Int J Biochem Cell Biol. 2018;99:91–9. doi: 10.1016/j.biocel.2018.04.005. [DOI] [PubMed] [Google Scholar]
- 58.Deng Y, Wang F, Hughes T, Yu J. FOXOs in cancer immunity: knowns and unknowns. Semin Cancer Biol. 2018;50:53–64. doi: 10.1016/j.semcancer.2018.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
HE histological staining and IHC staining of COX-2 in prostate tissues. Histological staining pictures (a–l) of prostate at the first day (a and d for BC; b and e for NS; c and f for CAR) and the 7th day (g and j for BC; h and k for NS; i and l for CAR) among the three groups are presented as above. Numerous infiltrated inflammatory cells have been found in prostatic stroma at the 7th day after carrageenan injection. Meanwhile, IHC images (m–x) of COX-2 at the first day (m and p for BC; n and q for NS; o and r for CAR) and the 7th day (s and v for BC; t and w for NS; u and x for CAR) are presented, the higher magnification (×400) and local representative visual area are indicated by white arrows as well as magnification in box. The expression of COX-2 was significantly increased in prostatic glandular epithelium and infiltrated cells in stroma. The scale bars are consistently 200 µm. COX-2: cyclooxygenase-2; BC: blank control group; NS: normal saline group; CAR: carrageenan group.
COX-2 expression in BC, NS, and CAR groups at day 1 and day 7. The mean densities of prostate tissues from three cases were presented at the top graph and summarized in the bottom table. Mean ± s.e.m, n = 3, #P > 0.05, **P < 0.01, ***P < 0.001, compared with the control of each group. COX-2: cyclooxygenase-2; BC: blank control group; NS: normal saline group; CAR: carrageenan group; s.e.m: standard error of the mean.
Increased IFN-γ in rats with carrageenan-induced CNP. Circulating IFN-γ levels in serum samples of BC, NS, and CAR groups on day 7 after model establishment. Mean ± s.e.m, n = 3, ***P < 0.001, compared with BC and NS groups. IFN-γ: interferon-γ; BC: blank control group; NS: normal saline group; CAR: carrageenan group; s.e.m: standard error of the mean; CNP: chronic nonbacterial prostatitis.
GO enrichment bar plot of target genes of differentially expressed miRNA. The top ten enriched GO functions (based on P values) in separated categories of BP, CC, and MF. The number at the end of each column represents the number of differentially expressed genes in the corresponding enriched GO function, which could reflect the gene distribution on certain GO functions. (a) CAR versus BC; (b) CAR versus NS. BC: blank control group; NS: normal saline group; CAR: carrageenan group; GO: gene Ontology; BP: biological process; CC: cellular component; MF: molecular function.
Relative expression of cAMP and mTOR in BC, NS, and CAR groups at day 7. Mean ± s.e.m., n = 3, **P < 0.01, compared with the control of each group. BC: blank control group; NS: normal saline group; CAR: carrageenan group; cAMP: cyclic adenosine monophosphate; mTOR: mammalian target of rapamycin; s.e.m: standard error of the mean.


