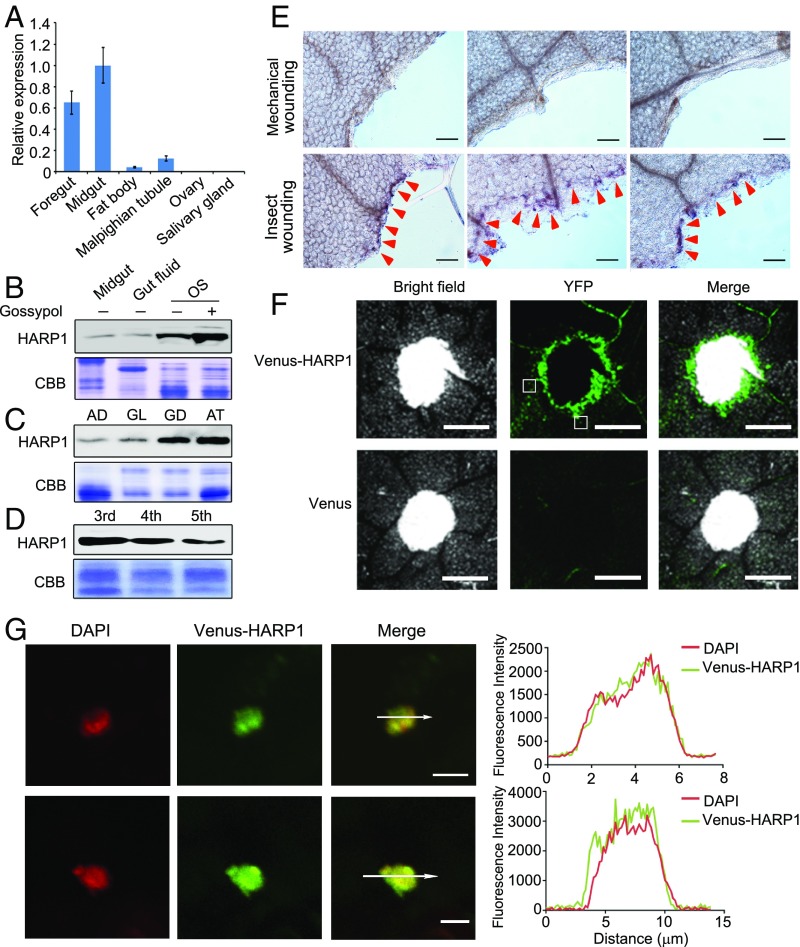
Fig. 1.
HARP1 in H. armigera OS migrates into the leaf cells through the wounding damage sites. (A) qRT-PCR analysis of HARP1 transcripts in indicated tissues of fifth-instar larvae. The expression level in midgut was set to 1. Error bars represent ± SD (n = 3 biological replicates). (B–D) Immunoblot detection of HARP1 protein level. The protein amount in each loading was quantified by Bradford assay and visualized by Coomassie Brilliant Blue (CBB) staining. All of the experiments were repeated at least two times, and the results were consistent. In B, the fourth instar larvae were fed on artificial diet supplemented with (+) or without (−) 0.1% gossypol for 1 d, and total proteins were collected from midgut, gut fluid and OS. In C, the OS was collected from the fourth instar larvae fed on artificial diet (AD), glandless (GL), or glanded (GD) cotton and A. thaliana (AT) leaves for 1 d. In D, the OS was collected from the indicated instar larvae that were fed on AT leaves for 1 d. (E) Whole amount immunohistochemistry detection of HARP1 at the chewing sites (red arrows) of Arabidopsis leaves. The mechanical wounding leaves were used as negative control. Anti-HARP1 antibody was used to detect HARP1 in B–E. (F and G) Translocation of the Venus-HARP1 fusion protein into plant cells through damage sites. The Arabidopsis leaves were punched and incubated with the protein solutions of Venus-HARP1 or Venus for 1 h and washed for three to four times to remove the extra proteins that adhered on the leaf surface. The boxes indicate the location as shown in G. (F) Venus-HARP1 but not Venus was detected at the wounded sites. (G) A portion of Venus-HARP1 was located in the nucleus of leaf cell. Fluorescence intensity in cross-section (white arrow) is shown. (Scale bars: E, 100 μm; F, 500 μm; G, 5 μm.)