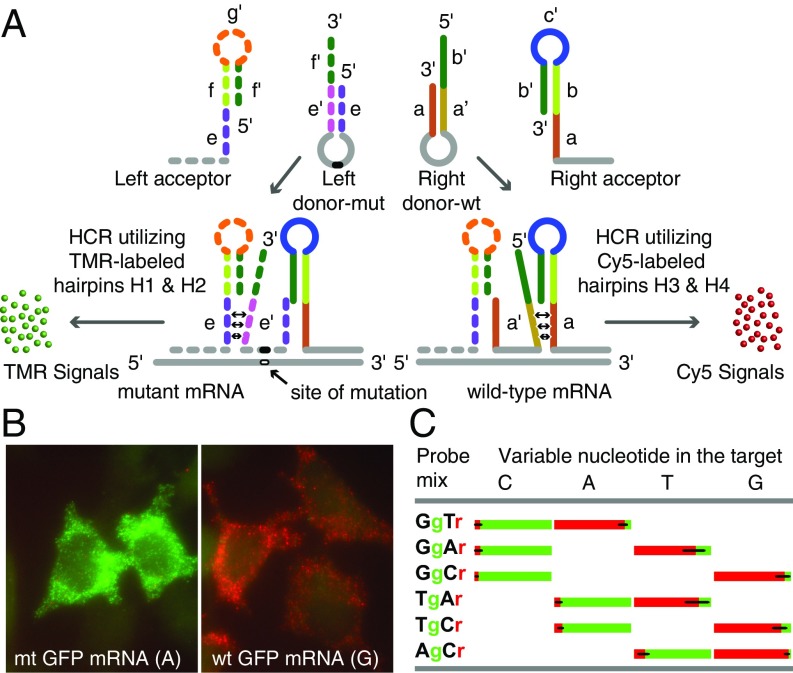
Fig. 2.
Generating amplified HCR signals from SNVs in single mRNA molecules using 4 interacting hairpin probes. (A) Depending on whether the target is mutant or wild-type, only one of the 2 donor hairpin probes binds to the target sequence in each mRNA, whereas both acceptor probes bind to it. The binding of a right donor-wt to a wild-type target sequence initiates a strand-displacement reaction in the right acceptor that leads to generation of a red HCR signal using Cy5-labeled HCR hairpins H3 and H4. The binding of a left donor-mut to a mutant target sequence initiates a strand-displacement reaction in the left acceptor that leads to generation of a green HCR signal using TMR-labeled HCR hairpins H1 and H2. (B) Examples of signals in either red or green channels after HCR. Cells were probed using a Tg-Cr probe mixture. This code identifies the discriminating nucleotide in the donor hairpin and lists the representative color of the HCR spots generated if that donor hairpin hybridizes to its target sequence. Cells expressing wild-type mRNAs yielded mostly red signals, and those expressing mutant mRNAs yielded mostly green signals. (C) Fractions of green and red spots from cells transfected with one of the 4 GFP variants that differed by the identity of the nucleotide (top) at one position. Each variant was probed using 3 pairwise combinations of the left and right donors (so that at least one donor was fully complementary to the target) together with common right and left acceptors. The bar graphs show the average percentage of spots seen in each color in single cells.