Fig. 3. The SRAP DPC forms a thiazolidine linkage stabilized by conserved residues.
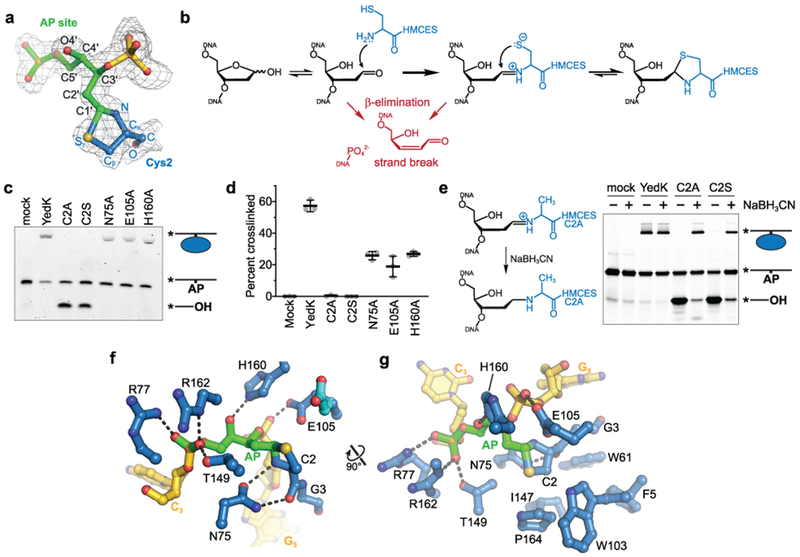
a-h, a, The DPC between the AP site (green) and Cys2 (blue) superimposed against 2Fo-Fc composite annealed omit electron density contoured at 1σ. b, Proposed chemical mechanism of the crosslinking reaction with competing lyase reactions in red. c, Representative denaturing PAGE gel showing crosslinking and lyase activity of YedK mutants. Bands were visualized by FAM fluorescence. d, Crosslinking efficiencies of YedK mutants (mean ± SD, n=3 independent measurements). e, NaBH3CN was added to crosslinking reactions to trap the Schiff base intermediates of YedK C2A and C2S mutants. The NaBH3CN-reduced Schiff base is refractory to β-elimination. Bands were visualized by FAM fluorescence. f, Residues contacting the DPC (DNA, gold; AP site green; protein, blue). The alternate Glu105 conformer is cyan. Dashed lines denote hydrogen bonds. g, Orthogonal view showing hydrophobic residues cradling Cys2. The second Glu105 conformer is not shown for clarity. Uncropped gel images are shown in Supplementary Data Set 1. Source data for d are available online.
