Abstract
Background: Pancreatic cancer is one of the most aggressive human malignancies that is associated with early metastasis and chemoresistance. Tropomyosin (TPM) is an indispensable regulatory protein for muscle contraction, Abnormal expressions of TPM gene are closely related to the carcinogenesis and metastasis of malignant tumors.
Purpose: In this experiment, a monoclonal stable transfected cell line was established by the knock-down of TMP3 expression in PANC-1 cells with the lentivirus method, and the impacts of the downregulated TPM3 gene expression on the EMT-related molecules and biological behaviors of PANC-1 cells were explored.
Methods: Based on the TPM3 gene sequence, we designed the RNA interference sequence, constructed and screened out the recombinant plasmid segment TPM3-shRNA with the optimal silencing effect, and carried out lentivirus titer determination and packaging. The recombinant lentiviral interference vector LV-TPM3-shRNA was transfected into PANC-1 cells; the transfection efficiency was then evaluated to screen out the monoclonal stable transfected PANC-1 cell line with downregulated TPM3 expression. The qRT-PCR and Western blot were used to detect the changes in the gene- and protein-levels expressions of EMT-related transcription factors in the target cell line and to respectively test the variations of the invasion and proliferation capacities.
Results: It is shown that the monoclonal stable transfected PANC-1 cell line with downregulated TPM3 expression was successfully established with the recombinant lentiviral vector. After knocking down the expression of TPM3 gene in PANC-1 cells, EMT occurred in the cells; the cell phenotype showed malignant transformation, and the in vitro biological behaviors of the cells (such as proliferation and invasion) were enhanced to different degrees.
Conclusion: It is indicated that the TPM3 gene can be a potential target spot for the treatment of pancreatic cancer.
Keywords: pancreatic cancer, TPM3, lentivirus, EMT, proliferation, invasion
Introduction
Pancreatic cancer is a highly malignant tumor of the digestive system and featured by concealed clinical manifestations, a high difficulty of early diagnosis, a high degree of malignancy, and an extremely low cure rate.1,2 The worldwide incidence and mortality of pancreatic cancer are almost the same, the incidence rate has increased year by year, and the mortality rate has been maintained at a very high level. Even with systematic treatment, the 5-year survival rate of pancreatic cancer is still <5%. In this sense, exploring and clarifying the molecular mechanisms of pancreatic cancer occurrence and progression as well as the characteristics of the invasion and metastasis processes can lay an experimental foundation and theoretical basis for the improvement of the early diagnosis rate and the determination of an effective therapeutic target spot, showing fundamental clinical significance.
Tropomyosin (TPM) is an indispensable regulatory protein for muscle contraction and is widely expressed in human cells. Under normal conditions,3 it maintains the stability of cytoskeletal structure and plays an important role in the processes, including cell movement, material transport, cell apoptosis, and signal transduction.4 Currently, the common TPMs in mammals involve TPM1, TPM2, TPM3, TPM4, TPM5, etc. and exist in various isomeric structures.5–8 Abnormal expressions of the TPM gene family are strongly associated with hypertrophic cardiomyopathy, myasthenia gravis, nerve tissue repair, and allergic reactions. It has been confirmed in recent studies that the abnormal expression of TPM gene initially leads to only morphological changes of the cytoskeleton, but ultimately causes tissue fibrosis and carcinogenesis. Abnormal expressions of TPM gene are closely related to the carcinogenesis and metastasis of malignant tumors.
The siRNA vectors adopted in current RNAi technologies suppress the expressions of target genes with specific sequences and, just like gene knockout technology, can only produce transient expression. However,9 the effective integration of a lentiviral vector with a TPM3 gene can effectively resist the resistance of the host cell, prevent the off-target effect, and stabilize the gene silencing effect. The lentiviral vector used in our experiments also carried an enhanced green fluorescent protein (GFP) gene, which is called a marker gene and has higher brightness and sharpness than average fluorescent proteins; by constructing a fusion integrating the expressions of a cloned TPM3 gene and a GFP gene,10 both in vitro and vivo experiments allow clear observation on whether the target gene is transfected into a PANC-1 cell line and the detection of the transfection efficiency.11 In this experimental study, the lentiviral vector shRNA was combined with the RNAi technology; the lentiviral vector was introduced exogenously into the cells, and the cells stably and continuously produced shRNA to efficiently block the expression of the target gene.
The epithelial-to-mesenchymal transition (EMT) is a potential metastasis mechanism in tumorigenesis and has currently been found in malignant tumors such as liver cancer, gastric cancer, pancreatic cancer, colorectal cancer, and breast cancer. The EMT is a dynamic changing process; the early stage is mainly featured by the downregulated expression of epithelial markers (E-cadherin, β-catenin, occludin, etc.) and the upregulated expression of interstitial markers (vimentin and fibronectin), leading to the loss of cell polarity and the reconstitution of the cytoskeleton. Under a pathological condition,12,13 the EMT causes tissue and organ fibrosis and enhances the activity range and invasive property of cells, making tumors easily migrate away from the primary focus and pass through the basilar membrane to invade and metastasize to the surrounding tissues. Moreover,14 the inflammatory environment under which the tumor cells are located also promotes the EMT, which enhances the antiapoptosis property of the cancer cells, resulting in a vicious circle.
In the earlier stage of our study, we had found that the expression of TPM3 was different in pancreatic cancer tissues and paracancerous tissues. Compared to normal tissue, the TPM3 has a lower expression in pancreatic cancer. Therefore, after the expression pattern of TPM3 in pancreatic cancer was confirmed, we focused on the establishment of LV-TPM3-shRNA and the monoclonal stable strains, and the expression of related genes in pancreatic cancer. In this experiment, the TPM3 gene expression was knocked down with the lentiviral interference vector; the level of E-cadherin (the epithelial phenotype marker) decreased, the expression of vimentin (the mesenchymal phenotype marker) increased, the EMT occurred in the cells, and the in vitro biological properties of pancreatic cancer cells (including invasion, proliferation, and migration) were enhanced. The preliminary study has shown that TPM3 gene is closely related to the occurrence of EMT and the metastasis of pancreatic cancer.
Materials and methods
Cell culture
A PANC-1 cell line sourced from the Chinese Academy of Sciences was cultured in 5 mL DMEM complete culture solution (Gibco, New York, NY, USA; containing 10% FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin). The cell line was incubated in a constant-temperature incubator (5% CO2; 37 °C). The cells in logarithmic growth phase were then inoculated into a 6-well plate 24 hrs before the transfection and then transfected with a lentiviral solution.
Establishment of lentiviral vector LV-TPM3-shRNA
The LV3 recombinant shuttle plasmid and the packaging plasmids pVSV-G, pRev, and pGag/Pol were established and prepared by GenePharma Company, and the primers and related transfection reagents were also purchased from GenePharma. Determine the effective target sequences for the TPM3 gene shRNA (Group 1: 5ʹ-GAGGTATGAAGGTTATTGAAA-3ʹ;Group 2: 5ʹ-GGCAGATAGGAAGTATGAAGA-3ʹ, and Group 3: 5ʹ-GGAGCACCTCTGTACACAAAG-3ʹ). The designed Oligo primer (the positive-sense strand: 5ʹ-GATCC-(GN18)-(TTCAAGAGA)-(N18C)-TTTTTTG-3ʹ;the antisense strand: 3ʹ-G(CN18)-(AAGTTCTCT)-(N18G)-AAAAAACTTAA-5ʹ) was complementary to the cohesive end formed after the BamHI enzyme and EcorRI enzyme digestion, and the LV3-shDNA template was annealed to form the double-stranded DNA. The annealed product was connected to the vector fragment processed with enzyme digestion linear treatment. The connection production was transformed into competent cell E. coli DH5a. The recombinant positive clones were selected for PCR identification. The identification suggested that the enzyme digestion result was consistent with the clone sequencing. A lentiviral packaging kit was used for lentiviral packaging, concentration, and titer determination. After gradient dilutions, the lentiviral solutions were used to infect TPM3 cells. The infection efficiencies were calculated and the establishment results were evaluated.
Transfection and screening of a stable PANC-1 cell line
The experimental cells were divided into five groups – the blank control sample group (the B-S group), the negative control sample group (the NC-S group), and cell infected with lentiviral interference vector sample groups (the 1/2/3-S groups, including LV-shTPM3-1-S, LV-shTPM3-2-S, and LV-shTPM3-3-S). Puromycin at gradient concentrations was added to every group. The groups were observed under a microscope after 7 days. The obtained minimum lethal concentration was used to screen out the lentivirus-infected PANC-1 cell line.
qRT-PCR analysis
The total RNA in every group was extracted with the TRIzol reagent and was treated with inverse transcription to prepare the cDNA. The Taq DNA polymerase and related PCR reagents were used for the amplification. Strictly following the instructions of the reagents, the TPM3 and GAPDH primers sequences were designed as follows: (TPM3 positive-sense strand: 5ʹ-GGAGACTTGGAACGCACAGA-3ʹ,antisense strand: 5ʹ-TTCAGCAGCACTCAGACACTTC-3ʹ;GAPDH positive-sense strand: 5ʹ-CATGAGAAGTATGACAACAGCCT-3ʹ, antisense strand: 5ʹ-AGTCCTTCCACGATACCAAAGT-3ʹ). A total of 3 duplicate wells were set for every group, and the reaction procedure was as follows – 95 °C, 3 mins, denaturation; 95 °C, 30 s; 62 °C, 40 s; 40 cycles. The GADPH fragment was used as the internal reference; the ΔΔCt analysis method was used to calculate the data obtained through the qPCR instrument; the 2−ΔΔCt method was utilized to calculate and compare the gene expressions in the groups and also applied to the identification of the TPM3 gene expressions in the stable transfected cell lines and the mRNA expressions in the subsequent EMT-related factors. The PCR primers for the EMT-related molecules were designed as summarized in Table 1.
Table 1.
The PCR primers for the EMT-related molecules
| Gene | Sequence (direction 5ʹ-3ʹ) | Amplification length(bp) |
|---|---|---|
| GAPDH | F-CATGAGAAGTATGACAACAGCCT | 113 |
| R-AGTCCTTCCACGATACCAAAGT | ||
| E-cadherin | F-CTTGGTCTACGCCTGGGACT | 130 |
| R-TGTGAGCAATTCTGCTTGGA | ||
| Vimentin | F-AATGGCTCGTCACCTTCGT | 160 |
| R-CAGATTAGTTTCCCTCAGGTTCA | ||
| Snail | F-CCTGGGTGCCCTCAAGAT | 263 |
| R-TGTGGAGCAGGGACATTCG | ||
| Twist | F-CGGGAGTCCGCAGTCTTAC | 161 |
| R-GCTTGAGGGTCTGAATCTTGCT |
Abbreviations: PCR, polymerase chain reaction; EMT, epithelial-to-mesenchymal transition.
Western blotting analysis
The transfected PANC-1 cells in every group were sequentially dissolved in the lysate; the total protein was extracted and separated with 12% SDS-PAGE, transferred to a micro-well plate of PVDF membrane for 1 hr, cultured in the TBST (containing 5% skimmed milk) under 37 °C for 2 hrs, and incubated overnight under 4 °C after the addition of the primary antibody (rabbit anti-human polyclonal TPM3 antibody,1:500, 33 kDa, ABclonal, A1206; rabbit anti-human polyclonal CDH1 antibody, 1:200, 135 kDa, ABclonal, A3044.rabbit anti-human polyclonal VIM antibody, 1:500, 54 kDa, ABclonal, A2666.mouse anti-human monoclonal GAPDH antibody, 1:10,000, Sigma, G8795.mouse anti-human monoclonal β-actin antibody, 1:10,000, Sigma, A5441). They went through membrane washing 3 times and incubated under 37 °C for 2 hrs after the addition of the secondary antibody (HRP-goat anti-rabbit IgG,1:10,000,JIR 111-035-003). After another membrane-washing, the ECL substrate was used for chemiluminescence test. Last, the Gel-pro-analyzer software was used to quantify the grayscale values and evaluate the transfection efficiencies. The method was also used to identify the expressions of TPM3 in the stable transfected cell lines and the subsequent EMT-related factors (E-cadherin and vimentin) at the protein level.According to the identification results of the stable screened cell lines, two groups of the experimental cell lines with the optimal transfection efficiencies (LV-shTPM3-1-S and LV-shTPM3-2-S) were selected from the stable transfected cell lines for the follow-up tests.
Invasion assay
Transwell chamber technology (Corning) for the detection of PANC-1 cell invasive property: According to the instructions of the reagent kit, detect the invasive property of each transfected group, take photos under the microscope, and take three 200× magnification fields of view per well for the photo-shooting; repeat the count of percentage of invasive cells for three times and calculate the average values, which were then used for statistical analysis.
CCK-8 assay
CCK-8 assay (Dojindo) for the detection of PANC-1 cell proliferation property: the cells in good growing condition 24 hrs after the transfection were prepared into cell suspensions, which were then paved in a 96-well plate. Use a ELIASA to detect the OD value at the wavelength of 450 nm and to obtain 6-hr data; the samples were measured and recorded at 4 hrs and 8 hrs, respectively. With the 6-hr data as the base points, the relative growth values were taken to draw growth curves.
Statistical analysis
All data in the experiments were measured repeatedly at least 3 times. The experimental data were statistically analyzed with SPSS 19.0 software. The measurement data were expressed as (x±s); data of multiple groups were compared with ANOVA analysis, while SNK-q test was used for inter-group comparison. The α=0.05 was taken as the level of significance, and p<0.05 suggested a statistically significant difference.
Results
Screening and establishment of the monoclonal recombinant lentivirus stable transfected PANC-1 cell lines
The results of the lentiviral Lv-NC-shRNA infection efficiencies (under 200× microscope): the negative control virus and culture solution were used successively to dilute the infection solution at the 1:10, 1:100, and 1:1000 concentrations (the virus titers were above 2×108 TU/mL); observe them under the microscope after 72 hrs (Figure 1A). The infection results showed that when the NC virus was infected for 72 hrs after the dilution with the culture medium at the rate of 1:10, the observed and calculated cell infection efficiencies were above 70%, suggesting the successful establishment of the recombinant lentiviral interference expression vector (Lv-TPM3-shRNA).
Figure 1.
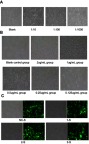
Screening and establishment of the monoclonal recombinant lentivirus-transfected PANC-1 cell lines.
Notes: (A) The cells were counted under the microscope after 1:10-, 1:100-, and 1:1000-gradient dilutions of the lentiviral solution; according to the dilution ratios, the virus titers were measured as above (2×108TU/mL), which met the packaging requirement and could infect PANC-1 cells. (B) In the experimental groups, puromycin of 2 µg/mL,1 µg/mL, 0.5 µg/mL, 0.25 µg/mL, and 0.125 µg/mL were added successively. The cells were observed under the microscope (200×) after 7 days. (C) Puromycin culture solutions were added to the above 4 groups, respectively. The cells were observed under the microscope after 7 days. The 1-S, 2-S, and 3-S groups represented the TPM3-infected lentiviral interference vector groups.
Abbreviations: 1:10, 1:10 gradient dilutions; 1:100, 1:100 gradient dilutions; 1:1000, 1:1000 gradient dilutions; TPM3, tropomyosin-3; NC-S, negative control sample; 1-S, LV-shTPM3-1-sample; 2-S, LV-shTPM3-2-sample; 3-S, LV-shTPM3-3-sample.
Results of the Exploration on the Minimum Lethal Concentration in the Stable Cell Line: The screening drug, puromycin, kills cells that do not express resistance genes. The puromycin culture solution was diluted to 0.125–2 µg/mL (Figure 1B). The observation on 7d confirmed that when the concentration of puromycin was 1 µg/mL, numerous cells died. Therefore, puromycin of 1 µg/mL was used for subsequent screening of monoclonal stable cell lines.The microscopic (200×) photos of the infected lentiviral stable cells: the lentivirus-transfected PANC-1 cell lines were screened with 1 µg/mL puromycin and then observed under an ordinary microscope and a fluorescence microscope. The fluorescent expression levels were homogeneous without loss propensity, and the cell infection efficiency reached about 90% (Figure 1C), demonstrating the successful construction of the monoclonal stable PANC-1 cell line.
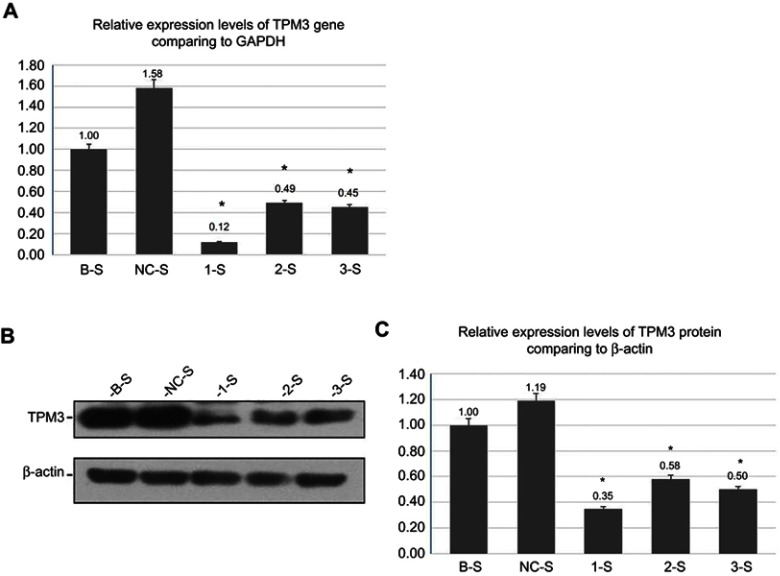
Identification of the monoclonal recombinant lentivirus-stable transfected PANC-1 cell lines
The results of the screened stable cell lines, are shown in Table 2 and Figure 2A. The electrophoresis of the stable transfected cell line RNA, TPM3, and GAPDH gene amplification products showed no nonpecific amplification. The 1-S group, the 2-S group, and the 3-S group were compared with the NC-S group, respectively, suggesting statistically significant differences (p<0.05). It was indicated that, after the screening of the monoclonal stable cell lines, the detection confirmed the downregulation of the TPM3 gene expression at the mRNA level and stable Lv-TPM3-shRNA interference effect at the gene level. But the NC-S group significantly differed from that of the B-S group (p<0.05) (Figure 2A). We analyzed this phenomenon in experiments that the empty vector virus solution maybe caused minimal damage or influence to PANC-1 cells in NC-S group, the other reason may be the expression of the TPM3 gene was unstable at the genetic level, which eventually leads to the statistically difference in the result.
Table 2.
Relative expression levels of TPM3 gene
| Groups | The relative expression of TPM3 mRNA 2−ΔΔCt |
|---|---|
| B-S | 1.00±0.05 |
| NC-S | 1.58±0.06 |
| 1-S | 0.12±0.00 * |
| 2-S | 0.49±0.02 * |
| 3-S | 0.45±0.01 * |
Notes: *Indicates that the 1-S group, the 2-S group, and the 3-S group were compared with the NC-S group, respectively (take the average of 3 measurements; the B-S group was used as the control).
Abbreviations: TPM3, tropomyosin-3; NC-S, negative control sample; 1-S, LV-shTPM3-1-sample; 2-S, LV-shTPM3-2-sample; 3-S, LV-shTPM3-3-sample.
Figure 2.
Identification of the monoclonal recombinant lentivirus-transfected PANC-1 cell lines.
Notes: (A) The qRT-PCR results for the relative expression levels of TPM3 gene: *indicates that the 1-S group, the 2-S group, and the 3-S group were compared with the NC-S group, respectively. (B) and (C) Gel figure of the relative expression levels of TPM3 protein and the histogram analysis results. *Indicates that the 1-S group, the 2-S group, and the 3-S group were compared with the NC-S group, respectively (take the average of 3 measurements; the B-S group was used as the control).
Abbreviations: TPM3, tropomyosin-3; B-S, blank control sample; NC-S, negative control sample; 1-S, LV-shTPM3-1-Sample; 2-S,LV-shTPM3-2-Sample; 3-S, LV-shTPM3-3-Sample.
Furthermore, we analyzed the expression level of TPM3 protein in the screened stable lines through the Western blot method, as shown in Figure 2B and C. Comparing the 1-S group, the 2-S group and the 3-S group with the NC-S group suggested statistically significant differences (p<0.05), while the comparison between the BS group and the NC-S group showed no statistical significance (p>0.05). It was found by the screening of the monoclonal stable transfected cell lines that the expression of the TPM3 gene was downregulated at the protein level, indicating that the lentiviral vector-mediated effect was confirmed by qRT-PCR and Western blot tests to be significant in the transiently transfected cell lines; the results after stable screening were consistent with the transient results; the expressions of the TPM3 gene in the PANC-1 cells were significantly downregulated and maintained at a stable level, suggesting the successful establishment of the monoclonal stable transfected PANC-1 cell line (Lv-TPM3-shRNA).
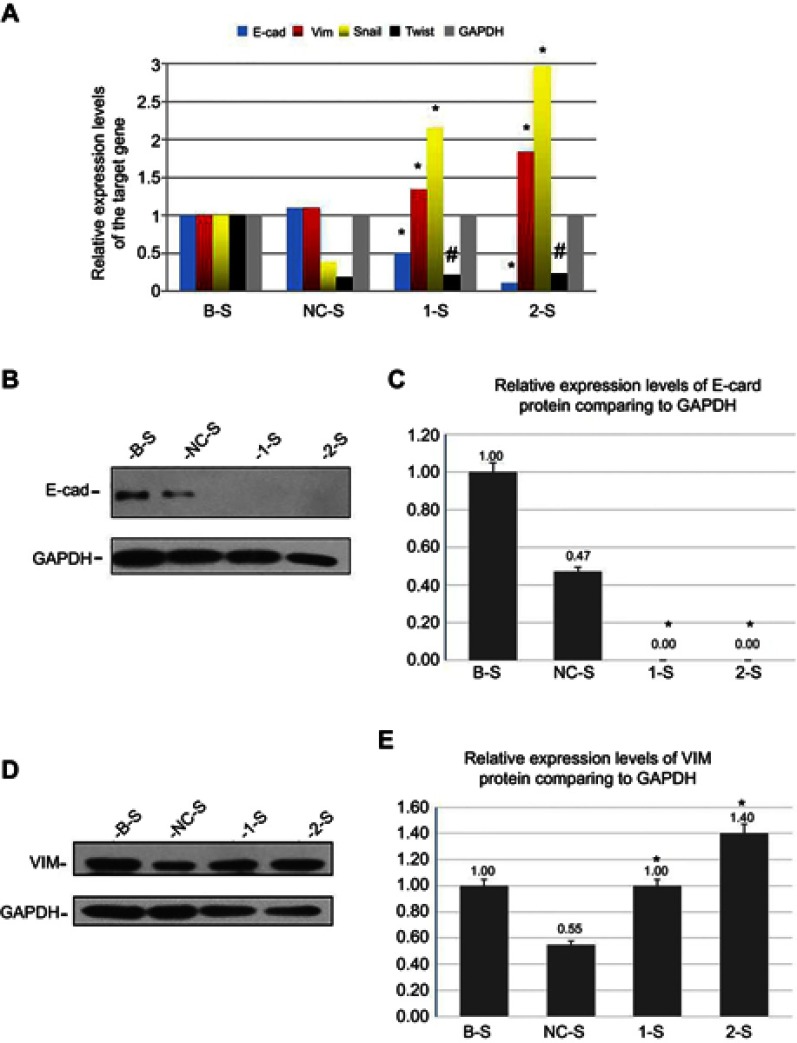
Detection of EMT-related genes in the monoclonal recombinant lentivirus-stable transfected PANC-1 cell lines
In the stable transfected cell lines, using the qRT-PCR method, the RNA electrophoresis of each related gene products showed no nonspecific amplification (Table 3, Figure 3A). The comparison of the 1-S group and the 2-S group with the NC-S group showed the downregulation of E-cadherin expression (p<0.05) and the upregulation of vimentin and Snail expressions (p<0.05). The expression of Twist gene was downregulated insignificantly (p>0.05), and the comparison between the B-S group and the NC-S group showed no statistically significant difference (p>0.05).
Table 3.
Relative quantification and expression levels of the target gene
| Groups | E-cad 2−ΔΔCt | Vim2−ΔΔCt | Snail 2−ΔΔCt | Twist 2−ΔΔCt |
|---|---|---|---|---|
| B-S | 1.00±0.04 # | 1.00±0.05 # | 1.00±0.07 # | 1.00±0.15 # |
| NC-S | 1.10±0.03 | 1.10±0.07 | 0.38±0.02 | 0.19±0.03 |
| 1-S | 0.51±0.02 * | 1.34±0.09 * | 2.15±0.16 * | 0.22±0.02 # |
| 2-S | 0.11±0.02 * | 1.84±0.09 * | 2.97±0.52 * | 0.24±0.03 # |
Notes: *Indicates that the 1-S group and the 2-S group were compared with the NC-S group, respectively, and showed a statistically significant difference (p<0.05). #Indicates that the difference is not statistically significant (p>0.05) (take the average of 3 measurements; the B-S group was used as the control).
Abbreviations: E-cad, E-cadherin; Vim, vimentin; NC-S, negative control sample; 1-S, LV-shTPM3-1-sample; 2-S, LV-shTPM3-2-sample.
Figure 3.
Detection of EMT-related genes in the monoclonal recombinant lentivirus-transfected PANC-1 cell lines.
Notes: (A) Histogram of the relative quantification and expression levels of the target gene (E-cadherin, vimentin, Snail, Twist, and GAPDH). *Indicates a statistically significant difference (p<0.05). #Indicates that the difference is not statistically significant (p>0.05). By using Western blot,the gel figure of the relative expression levels of E-cadherin (B and C) and vimentin (D and E) proteins and the histogram analysis results of the stable transfected cell lines. *Indicates a statistically significant difference (p<0.05) (every data was repeatedly measured 3 times,take the average of 3 measurements; the B-S group was used as the control).
Abbreviations: E-cad, E-cadherin; VIM, vimentin; 1-S, LV-shTPM3-1-sample; 2-S, LV-shTPM3-2-sample.
Furthermore, using Western blot method, we analyzed the results of the stable transfected cell lines, as shown in Figure 3B-E. When the 1-S group and the 2-S group were compared with the NC-S group, the expressions of the epithelial phenotype marker E-cadherin proteins were downregulated (p<0.05) (Figure 3B and C), while the expressions of the mesenchymal phenotype marker vimentin proteins were upregulated (p<0.05) (Figure 3D and E). The EMT molecular markers changed, and EMT phenotypic transformation occurred in cells at the molecular level. But the comparison between the B-S group and the NC-S group showed a statistically significant difference (p<0.05) (Figure 3C and E).
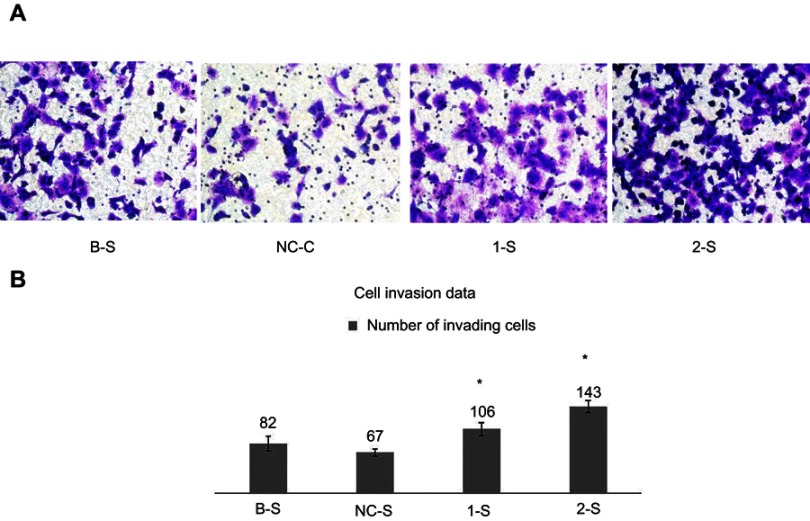
Transwell chamber technology for the determination of the PANC-1 cell invasive properties
The counts of invading cells in the experimental groups were significantly higher than that in the NC-S group (p<0.05), while the comparison between the B-S group and the NC-S group suggested no significant difference (p>0.05) (Figure 4A and B). It was suggested that the invasive property of the PANC-1 cells was enhanced significantly after the downregulation of TPM3 gene expression.
Figure 4.
Transwell chamber technology for the determination of the PANC-1 cell invasive properties.
Notes: (A) and (B) Analysis of the cell invasion data: in every group, a total of 5 different view fields were used to count the numbers of the invading cells. *Indicates that the 1-S group and the 2-S group were compared with the NC-S group, and the count was repeated 3 times.
Abbreviations: B-S, blank control sample; NC-S, negative control sample; 1-S, LV-shTPM3-1-sample; 2-S, LV-shTPM3-2-sample.
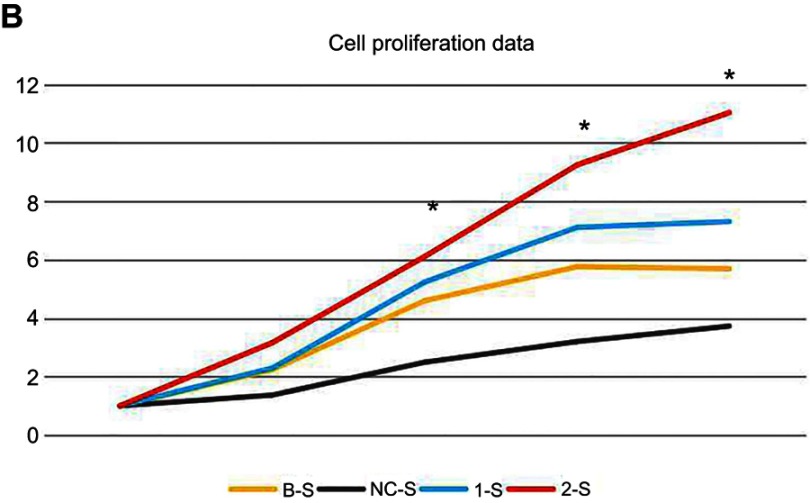
CCK-8 method for the determination of PANC-1 cell proliferation property
At the 24-hr/48-hr/72-hr/96-hr time points, the comparisons of the 1-S group and the 2-S group with the NC-S group both showed significant differences (p<0.05) (Table 4), suggesting significantly higher cell proliferation properties than the B-S group and the NC-S group after 24 hrs; the proliferation properties of the PANC-1 cells were enhanced after the downregulation of TMP3 expression (Figure 5A); however, the comparison between the B-S group and the NC-S group suggested no statistically significant difference (p>0.05).
Table 4.
Cell proliferation data
| The relative growth value | B-S | NC-S | 1-S | 2-S |
|---|---|---|---|---|
| 6 hrs/6 hrs | 1.00±0.02 | 1.00±0.03 | 1.00±0.02 | 1.00±0.01 |
| 24 hrs/6hrs | 2.23±0.01 | 1.37±0.04 | 2.29±0.03 * | 3.16±0.01 * |
| 48 hrs/6 hrs | 4.60±0.03 | 2.49±0.02 | 5.24±0.04 * | 6.11±0.03 * |
| 72 hrs/6 hrs | 5.76±0.03 | 3.20±0.03 | 7.11±0.04 * | 9.25±0.06 * |
| 96 hrs/6 hrs | 5.69±0.04 | 3.73±0.04 | 7.31±0.01 * | 11.04±0.02 * |
Notes: *Indicates that the 1-S group and the 2-S group were compared with the NC-S group, respectively, and showed a statistically significant difference (p<0.05) (take the average of 3 measurements; the B-S group was used as the control).
Abbreviations: B-S, blank control group; NC-S, negative control sample; 1-S, LV-shTPM3-1-sample; 2-S, LV-shTPM3-2-sample.
Figure 5.
CCK-8 method for the determination of PANC-1 cell proliferation property.
Notes: (A) and (B) Analysis of the cell proliferation data: the cell proliferation was analyzed in the five groups to obtain cell relative growth data. *Indicates that the 1-S group and the 2-S group were compared with the NC-S group at the 24-hr/48-hr/72-hr/96-hr time points, and the measurements at each time point was repeated 3 times.
Abbreviations: B-S, blank control sample; NC-S, negative control sample; 1-S, LV-shTPM3-1-sample; 2-S, LV-shTPM3-2-sample.
Discussion
Currently, there is no research report on TPM3 (TPM) gene in pancreatic cancer, but TPM3 gene is abnormally expressed in many tumor cells. For example, the expression of TPM3 is upregulated in15 liver cancer,16 thyroid cancer, and17 ovarian cancer, but downregulated in18 colon cancer,19,20 breast cancer, and21 esophageal cancer. However, the results are inconsistent in studies conducted by different researchers and studies on different tumors.These findings suggest strong relationships between abnormal expressions of TPM gene and tumorigenesis.
When constructing the lentiviral interference vector (Lv-TPM3-shRNA), we found that after transient transfection of the recombinant vector into the PANC-1 cells, although the detection confirmed the downregulated TPM3 gene expression, the fluorescent expression levels of the TPM3 genes in the cells were not uniform; besides, there was a high possibility of fluorescence loss as the extension of the screening time. After the drug resistance screening, the cell fluorescence intensities of the stable transformed cell lines were uniform, there was no fluorescence loss, and the expression of the target gene was steadily downregulated, suggesting the successful establishment of the monoclonal stable transfected PANC-1 cell lines.22,23 It was confirmed that the method of using lentivirus to establish a TPM3 monoclonal stable cell line in PANC-1 cells is feasible and superior, providing a theoretical basis for subsequent research.
E-cadherin has long been regarded as a tumor metastasis suppressor gene as well as a key gene in the EMT process, and its abnormal expression is the molecular basis of cell dissociation.24 It has been found that a tumor with upregulated E-cadherin expression is featured by a high degree of tumor differentiation, slow growth, lower metastasis propensity, and better prognosis, while the expression of E-cadherin is downregulated more significantly in tumors with higher malignancy degree and poorer differentiation.25–28 Many studies have shown that Snail, ZEB, KLF8, and other genes can directly suppress the expression of E-cadherin at the transcriptional level, while Twist and FoxC2 play a suppressing role indirectly.29,30 In vertebrates, suppressing the promoter of E-cadherin at the apparent level by DNA hypermethylation can silence the tumor-suppressor gene, induce the expressions of the relevant differentiation genes, and generate the EMT process,31 and the EMT cells induced in this way are more stable in the biological behaviors. Therefore, the downregulation of E-cadherin expression promotes the occurrence of the EMT and canceration of cells. Vimentin is a type III intermediate filament protein that is only expressed in mesenchymal cells and,32–34 under normal conditions, participates in the processes like forming integral cytoskeleton, repair of tissue damage, cell growth and apoptosis, and intercellular signal transduction. Vimentin is hardly expressed in epithelial-derived cells. When the EMT change occurs in cells, E-cadherin gene expression is downregulated, while vimentin expression is upregulated; the two are negatively correlated and lead to decreased intercell adhesion and enhanced cell migration and invasion. It is found in this study that the expressions of vimentin at both the gene and protein levels were significantly upregulated with the downregulation of TPM3 expression (p<0.05), and the monoclonal stable transfected PANC-1 cell line showed the interstitial cell morphology. It has been demonstrated that35 in breast cancer,36 liver cancer,37 gastric cancer, and38 prostate cancer, the upregulated expression of vimentin enhances the proliferation, invasion, and migration of cancer cells;39 in colon cancer, the downregulation of vimentin gene expression might lower the adhesive and invasive properties of the cancer cells.During the study process, it was found that the expressions of EMT-related transcription factors (such as Snail) were upregulated after the TPM3 gene downregulation (p<0.05), but the Twist expression was downregulated in an insignificant way (p>0.05). Numerous studies have shown that the transcription factor Snail can bind to the E-box of the E-cadherin promoter (the core sequence is CAGGTG), downregulating the expression of E-cadherin and inducing EMT. We also demonstrated with transwell technology and CCK-8 tests in the in vitro experiments that, comparing with the control group, the invasion and proliferation abilities of the PANC-1 cells were significantly enhanced (p<0.05), while the cell morphology also transited from epithelial to mesenchymal cell morphology. In summary, it is further convinced by the study on cell biological behaviors that the downregulation of TPM3 gene expression results in the abnormal activation of E-cadherin and vimentin genes, leading to the morphological changes of the pancreatic cancer cells, increasing the malignancy, facilitating the metastasis of pancreatic cancer via EMT, and making the cure rather more difficult.
In conclusion, by using the recombinant lentiviral vector,we successfully established a monoclonal stable PANC-1 cell line with downregulated TPM3 gene expression for the first time. Besides, it is demonstrated in the experiment that the knockdown of TPM3 gene expression promotes the occurrence and development of pancreatic cancer; the pathogenesis is further explored. Furthermore, to obtain more information about the expression of TPM3 in different pancreatic cancer cell lines, we will select more pancreatic cancer cell lines to study at the same time in the follow-up in-depth study. However, the study has laid an experimental foundation and theoretical basis for the early diagnosis of pancreatic cancer and for the determination of a therapeutic target spot.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel R, Miller K, Jemal A. Cancer statistics. CA Cancer J Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332 [DOI] [PubMed] [Google Scholar]
- 2.Mettu NB, Abbruzzese JL. Clinical insights into the biology and treatment of pancreatic cancer. J Oncol Pract. 2016;12(1):17–23. doi: 10.1200/JOP.2015.009092 [DOI] [PubMed] [Google Scholar]
- 3.Khaitlina SY. Tropomyosin as a regulator of actin dynamics. Int Rev Cell Mol Biol. 2015;318:255–291. doi: 10.1016/bs.ircmb.2015.06.002 [DOI] [PubMed] [Google Scholar]
- 4.Lees JG, Bach CT, O’Neill GM. Interior decoration: tropomyosin in actin dynamics and cell migration. Cell Adh Migr. 2011;5(2):181–186. doi: 10.4161/cam.5.2.14438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thierfelder L, Watldns H, MacRae C, et al. α-tropomyosin and cardiac troponin T mutations cause familial hypertrophic cardiomyopathy: A disease of the sarcomere. Cell. 1994;77:701–712. [DOI] [PubMed] [Google Scholar]
- 6.Malfatti E, Schaeffer U, Chapon F, et al. Combined cap disease and nemaline myopathy in the same patient caused by an autosomal dominant mutation in the TPM3 gene. Neuromuscul Disord. 2013;23(12):992–997. doi: 10.1016/j.nmd.2013.07.003 [DOI] [PubMed] [Google Scholar]
- 7.Guven K, Gunning P, Fath T. TPM3 and TPM4 gene products segregate to the postsynaptic region of central nervous system synapses. Bioarchitecture. 2011;1(6):284–289. doi: 10.4161/bioa.1.6.19336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gamez C, Sanchez-Garcia S, Ibanez MD, et al. Tropomyosin IgE-positive results are a good predictor of shrimp allergy. Allergy. 2011;66(10):1375–1383. doi: 10.1111/j.1398-9995.2011.02663.x [DOI] [PubMed] [Google Scholar]
- 9.Naldini L, Trono D, Verma IM. Lentiviral vectors,two decades later. Science. 2016;353(6304):1101–1102. doi: 10.1126/science.aah6192 [DOI] [PubMed] [Google Scholar]
- 10.Krinner S, Heitzer A, Asbach B, et al. Interplay of promoter usage and intragenic CpG content: impact on GFP reporter gene expression. Hum Gene Ther. 2015;26(12):826–840. doi: 10.1089/hum.2015.075 [DOI] [PubMed] [Google Scholar]
- 11.Li M, Rossi JJ. Lentiviral vector delivery of siRNA and sh RNA encoding genes into cultured and primary hematopoietic cell. Methods Mol Biol. 2008;433:281–299. [DOI] [PubMed] [Google Scholar]
- 12.Thiery JP, Lim CT. Tumor dissemination: an EMT affair. Cancer Cell. 2013;23(3):272–273. doi: 10.1016/j.ccr.2013.03.004 [DOI] [PubMed] [Google Scholar]
- 13.Diepenbruck M, Christofori G. Epithelial-mesenchymal transition(EMT) and metastasis: yes, no, maybe? Curr Opin Cell Biol. 2016;43:7–13. doi: 10.1016/j.ceb.2016.06.002 [DOI] [PubMed] [Google Scholar]
- 14.Kuilman T, Michaloglou C, Vredeveld LC, et al. Oncogene-induced senescence relayed by an interleuk independent inflammatory network. Cell. 2008;133(6):1019–1031. doi: 10.1016/j.cell.2008.03.039 [DOI] [PubMed] [Google Scholar]
- 15.Choi HS, Yim SH, Xu HD, et al. Tropomyosin3 overexpression and a potential link to epithelial-mesenchymal transition in human hepatocellular carcinoma. BMC Cancer. 2010;10:122. doi: 10.1186/1471-2407-10-663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greco A, Miranda C, Pierotti MA. Rearrangements of NTRK1 gene in papillary thyroid carcinoma. Mol Cell Endocrinol. 2010;321(1):44–49. doi: 10.1016/j.mce.2009.10.009 [DOI] [PubMed] [Google Scholar]
- 17.Tang HY, Lynn A. Protein isoform-specific validation defines multiple chloride intracellular channel and tropomyosin isoforms as serological biomarkers of ovarian cancer. J Proteomics. 2013;89:165–178. doi: 10.1016/j.jprot.2013.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mlakar V. Presence of activating KRAS mutations correlates significantly with expression of tumour suppressor genes DCN and TPM1 in colorectal cancer. BMC Cancer. 2009;9:282. doi: 10.1186/1471-2407-9-282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dube S, Yalamanchili S, Lachant J. et al. Expression of tropomyosin 1 gene isoforms in human breast cancer cell lines. Int J Breast Cancer;2015. 1–11. doi: 10.1155/2015/859427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Franzen B, Linder S, Uryu K, et al. Expression of tropomyosin isoforms in benign and malignant human breast lesions. Br J Cancer. 1996;73(7):909–913. doi: 10.1038/bjc.1996.162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zare M. Downregulation of tropomyosin-1 in squamous cell carcinoma of esophagus,the role of Ras signaling and ethylation. Mol Carcinog. 2012;51(10):796–806. doi: 10.1002/mc.20847 [DOI] [PubMed] [Google Scholar]
- 22.Hanoun N, Gayral M, Pointreau A, et al. Initial characterization of integrase- defective lentiviral vectors for pancreatic cancer gene therapy. Hum Gene Ther. 2016;27(2):184–192. doi: 10.1089/hum.2015.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ravet E, Lulka H, Gross F, et al. Using lentiviral vectors for efficient pancreatic cancer gene therapy. Cancer Gene Ther. 2010;17(5):315–324. doi: 10.1038/cgt.2009.79 [DOI] [PubMed] [Google Scholar]
- 24.Tiwari N, Gheldof A, Tatari M, et al. EMT as the ultimate survival mechanism of cancer cells. Semin Cancer Biol. 2012;22(3):194–207. doi: 10.1016/j.semcancer.2012.02.013 [DOI] [PubMed] [Google Scholar]
- 25.Galvan JA, Zlobec I, Wartenberg M, et al. Expression of E-cadherin repressors SNAIL,ZEB1 and ZEB2 by tumour and stromal cells influences tumour-budding phenotype and suggests heterogeneity of stromal cells in pancreatic cancer. Br J Cancer. 2015;112(12):1944–1950. doi: 10.1038/bjc.2015.177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang H, Liu L, Wang Y, et al. KLF8 involves in TGF-beta-induced EMT and promotes invasion and migration in gastric cancer cells. J Cancer Res Clin Oncol. 2013;139(6):1033–1042. doi: 10.1007/s00432-012-1363-3 [DOI] [PubMed] [Google Scholar]
- 27.Zhao J, Dong D, Sun L, et al. Prognostic significance of the epithelial-to- mesenchymal transition markers e-cadherin,vimentin and twist in bladder cancer. Int Braz J Urol. 2014;40(2):179–189. doi: 10.1590/S1677-5538.IBJU.2014.02.07 [DOI] [PubMed] [Google Scholar]
- 28.Jiang W, Fan H, Qian C, et al. Prognostic value of high FoxC2 expression in resectable non-small cell lung cancer, alone or in combination with E-cadherin expression. BMC Cancer. 2016;16:16–25. doi: 10.1186/s12885-016-2056-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang SQ, Zhang GQ. Correlation between methylation of the E-Cadherin gene and malignancy of prostate cancer. Genet Mol Res. 2016;15(2):1–9. [DOI] [PubMed] [Google Scholar]
- 30.Lombaerts M, Van Wezel T, Philippo K, et al. E-cadherin transcriptional down- regulation by promoter methylation but not mutation is related to epithelial-to- mesenchymal transition in breast cancer celllines. Br J Cancer. 2006;94:661. doi: 10.1038/sj.bjc.6602996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dumont N, Wilson MB, Crawford Y, et al. Sustained induction of epithelial to mesenchymal transition activates DNA methylation of genes silenced in basal-like breast cancers. Proc Natl Acad Sci. 2008;105(39):14867–14872. doi: 10.1073/pnas.0807146105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Menko AS, Bleaken BM, Libowitz AA, et al. A central role for vimentin in regulating repair function during healing of the lens epithelium. Mol Biol Cell. 2014;25(6):776–790. doi: 10.1091/mbc.E12-12-0900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mor Vaknin N, Punturieri A, Sitwala K, et al. Vimentin is secreted by activated macrophages. Nat Cell Bio. 2003;5(1):59–63. doi: 10.1038/ncb898 [DOI] [PubMed] [Google Scholar]
- 34.Yi EY, Park SY, Jung SY, et al. Mitochondrial dysfunction induces EMT through the TGF-β/Smad/Snail signaling pathway in Hep3B hepatocellular carcinoma cells. Int J Oncol. 2015;47(5):1845–1853. doi: 10.3892/ijo.2015.3154 [DOI] [PubMed] [Google Scholar]
- 35.Skalamera D, Dahmer-Heath M, Stevenson AJ, et al. Genome-wide gain of function screen for genes that induce epithelial-to-mesenchymal transition in breast cancer. Oncotarget. 2016;7(38):61000–61020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salvi A, Bongarzone I, Ferrari L, et al. Molecular characterization of LASP-1 expression reveals vimentin as its new partner in human hepatocellular carcinoma cells. Int J Oncol. 2015;46(5):1901–1912. doi: 10.3892/ijo.2015.2923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cong H, Yao RY, Sun ZQ, et al. DNA hypermethylation of the vimentin gene inversely correlates with vimentin expression in intestinal- and diffuse-type gastric cancer. Oncol Lett. 2016;11(1):842–848. doi: 10.3892/ol.2015.3937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lindsay CR, Le Moulec S, Billiot F, et al. Vimentin and Ki67 expression in circulating tumour cells derived from castrate-resistant prostate cancer. BMC Cancer. 2016;16:168–179. doi: 10.1186/s12885-016-2192-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mc Inroy L, Maatta A. Down-regulation of vimentin expression inhibits carcinoma cell migration and adhesion. Biochem Biophys Res Commun. 2007;360(1):109–114. doi: 10.1016/j.bbrc.2007.06.036 [DOI] [PubMed] [Google Scholar]