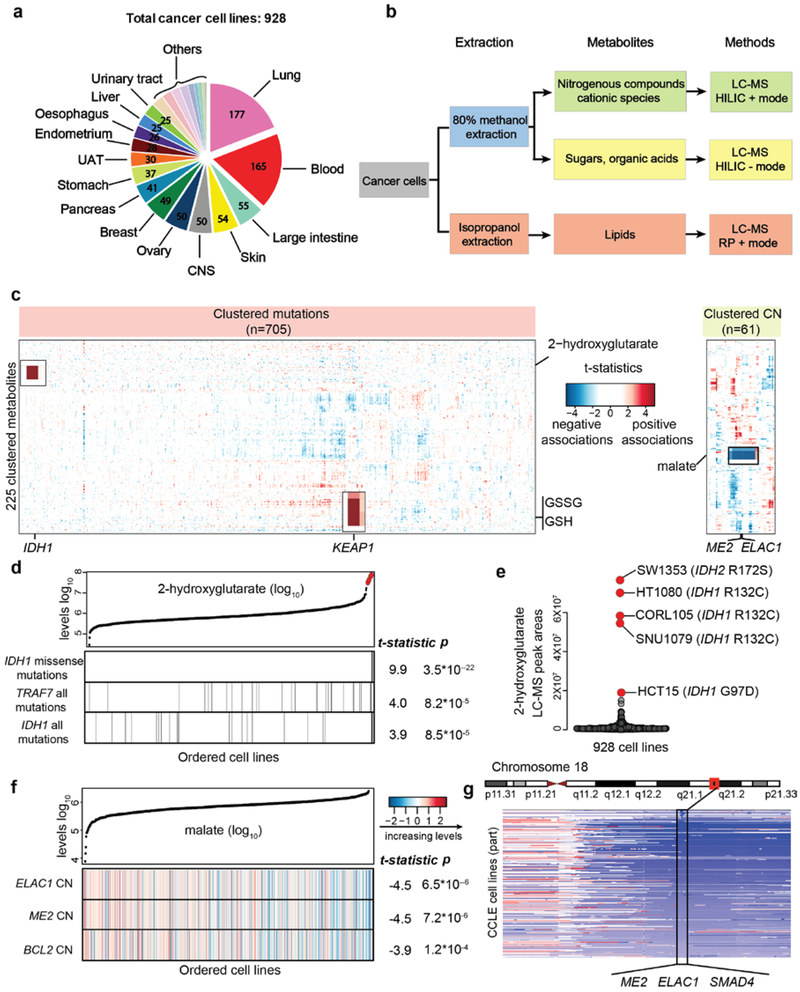
Fig.1 ∣. The CCLE database enables quantitative metabolomic modeling in relation to genetic features.
a, 928 cancer cell lines from more than 20 major tissues of origin were profiled for the abundance of 225 metabolites. The number of cell lines is annotated based on the tissues of origin. b, Schematic summarizing the workflow of metabolite profiling. c, Heatmap of 225 clustered metabolites (Y axis) and their associations with selected genetic features (X axis). T-statistics were calculated based on linear regression for each metabolite paired with each feature across all cell lines conditioned on the major lineages and were used to represent the regression coefficients scaled by standard deviations. Examples mentioned in the text are magnified and shown in black boxes. CN, copy number. d, 2HG and the top correlated mutations among all mutational features. Cell lines are shown as lines and ordered by increasing levels of 2HG. Those cell lines with corresponding mutations are labeled as black. The reported test statistics and p-values are based on the significance tests of genetic feature regression coefficients (cell line n=927, two-sided t-tests). e, Cancer cell lines with outlier levels of 2HG have specific IDH1/2 mutations. f, Malate levels and a heatmap representation of top correlated copy number alterations among all copy number features. The features are color-coded to indicate various copy number alterations (log2 scale) and are standardized for plotting. The reported test statistics and p-values are based on the significance tests of genetic feature regression coefficients (cell line n=912, two-sided t-tests). g, Schematic of the genomic locus containing ME2, ELAC1, and SMAD4.