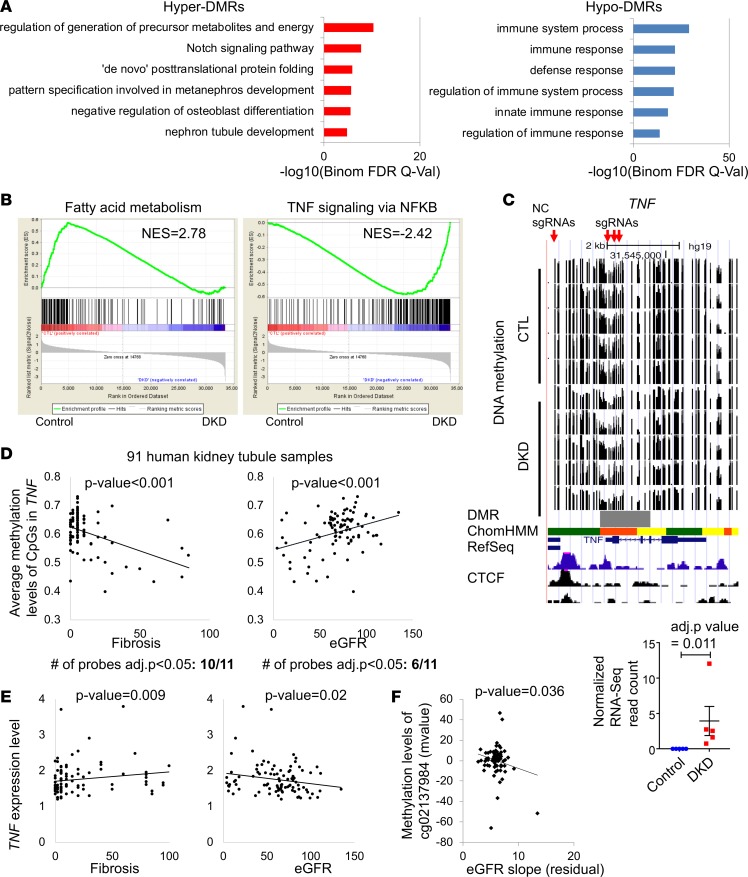
Figure 4. Increased enrichment for TNF pathway in human DKD tubules.
(A) Functional annotation analysis by GREAT of the hyper- and hypo-DMRs. (B) Gene set enrichment analysis (GSEA) results of genes that are increased and decreased in DKD. (C) WGBS DNA methylation level (control and DKD) followed by DMR highlight (gray), human kidney H3K27ac ChIP-seq, ChromHMM data, and RNA expression (by RNA sequencing). ChromHMM color code is from Figure 1C. (D) Correlation between GFR or interstitial fibrosis and CpG methylation in TNF gene locus measured by Infinium array in microdissected tubule samples from 91 human kidneys. (E) Correlation between eGFR or interstitial fibrosis and human patient TNF gene expression. P value was calculated using linear regression models adjusted for age, sex, race, diabetes mellitus, and hypertension (36). (F) Correlation between eGFR slope and the methylation of cg02137984 (Infinium array) which is localized in the TNF gene locus (31). P value was calculated using linear regression models adjusted for age, sex, race, diabetes mellitus, and hypertension.