Abstract
BACKGROUND
Recessive dystrophic epidermolysis bullosa (RDEB) is a severe form of skin fragility disorder due to mutations in COL7A1 encoding basement membrane type VII collagen (C7), the main constituent of anchoring fibrils (AFs) in skin. We developed a self-inactivating lentiviral platform encoding a codon-optimized COL7A1 cDNA under the control of a human phosphoglycerate kinase promoter for phase I evaluation.
METHODS
In this single-center, open-label phase I trial, 4 adults with RDEB each received 3 intradermal injections (~1 × 106 cells/cm2 of intact skin) of COL7A1-modified autologous fibroblasts and were followed up for 12 months. The primary outcome was safety, including autoimmune reactions against recombinant C7. Secondary outcomes included C7 expression, AF morphology, and presence of transgene in the injected skin.
RESULTS
Gene-modified fibroblasts were well tolerated, without serious adverse reactions or autoimmune reactions against recombinant C7. Regarding efficacy, there was a significant (P < 0.05) 1.26-fold to 26.10-fold increase in C7 mean fluorescence intensity in the injected skin compared with noninjected skin in 3 of 4 subjects, with a sustained increase up to 12 months in 2 of 4 subjects. The presence of transgene (codon-optimized COL7A1 cDNA) was demonstrated in the injected skin at month 12 in 1 subject, but no new mature AFs were detected.
CONCLUSION
To our knowledge, this is the first human study demonstrating safety and potential efficacy of lentiviral fibroblast gene therapy with the presence of COL7A1 transgene and subsequent C7 restoration in vivo in treated skin at 1 year after gene therapy. These data provide a rationale for phase II studies for further clinical evaluation.
TRIAL REGISTRATION
ClincalTrials.gov NCT02493816.
FUNDING
Cure EB, Dystrophic Epidermolysis Bullosa Research Association (UK), UK NIHR Biomedical Research Centre at Guy’s and St Thomas’ NHS Foundation Trust and King’s College London, and Fondation René Touraine Short-Exchange Award.
Keywords: Dermatology, Genetics
Keywords: Gene therapy, Genetic diseases, Skin
The LENTICOL-F trial is a human phase I study demonstrating safety and early efficacy of ex vivo lentiviral fibroblast gene therapy for adults with recessive dystrophic epidermolysis bullosa.
Introduction
Recessive dystrophic epidermolysis bullosa (RDEB) is one of the most severe forms of inherited skin fragility disorders, characterized by painful blisters and erosions leading to cutaneous infections, mutilating scarring, and aggressive cutaneous squamous cell carcinoma during early adulthood alongside other systemic complications (1). The most accurate incidence and prevalence of RDEB, estimated in 2002, were 3.05 per million live births and 1.35 per million population, respectively (2). RDEB is caused by biallelic loss-of-function mutations in COL7A1, encoding type VII collagen (C7) (3). C7 is the main component of anchoring fibrils (AFs) that ensure adherence of the epidermis to the dermis within the basement membrane. C7 is synthesized by both basal keratinocytes and dermal fibroblasts and consists of a central collagenous triple helix flanked by a large 145-kDa amino-terminal noncollagenous domain (NC1) and a smaller, 34-kDa carboxyl terminal noncollagenous domain (NC2) (4). In RDEB, malformed, deficient, or absent C7 compromises AFs and results in sublamina densa blisters.
RDEB has a major health economic burden; wound dressings for a 10-year-old child in the United States can cost $680 per day (5), which equates to more than $250,000 annually. Treatment options for RDEB are limited, particularly for correcting the underlying basement membrane C7 deficiency. Various therapeutic approaches under development include direct administration of recombinant C7 protein (6–8), hematopoietic stem cell transplantation (9, 10), infusion of allogeneic mesenchymal stromal cells (MSCs) (11, 12), MSC-derived exosomes (13), premature termination codon–readthrough (PTC-readthrough) drugs (14), and gene therapies (15–18). Among these strategies, one that promises potential correction of underlying C7 deficiency is ex vivo autologous gene-modified cell therapy using viral vectors. Retroviral vectors have been widely employed, achieving some efficacy and safety, particularly with the advent of self-inactivating (SIN) viral platforms (for review, see ref. 19). With respect to the cell target for gene therapy, although both keratinocytes and fibroblasts are involved in the production and secretion of C7, fibroblasts are generally more robust and easier to maintain in culture while allowing for a mode of delivery in the form of intradermal injections that will circumvent the clinical risk of skin graft failure and infections (20, 21). This rationale led us to develop a SIN lentiviral (LV) vector platform carrying a full-length codon-optimized COL7A1 cDNA under the control of the human phosphoglycerate kinase (PGK) promoter (Figure 1A) for the engineering of autologous RDEB fibroblasts for human application. Our preclinical assessment of this construct in an immunodeficient xenograft mouse model demonstrated C7 and AF restoration at the dermal-epidermal junction (DEJ) (22), and the same LV configuration has been used for phase I testing in adults with Netherton syndrome (ClinicalTrials.gov NCT01545323) (23). In light of the promising preclinical data (22), we conducted a phase I, open-label, single-center clinical trial evaluating the safety and early efficacy of SIN LV–mediated ex vivo gene-modified autologous fibroblast therapy in 4 adults with RDEB over 12 months (Figure 1B and Figure 2).
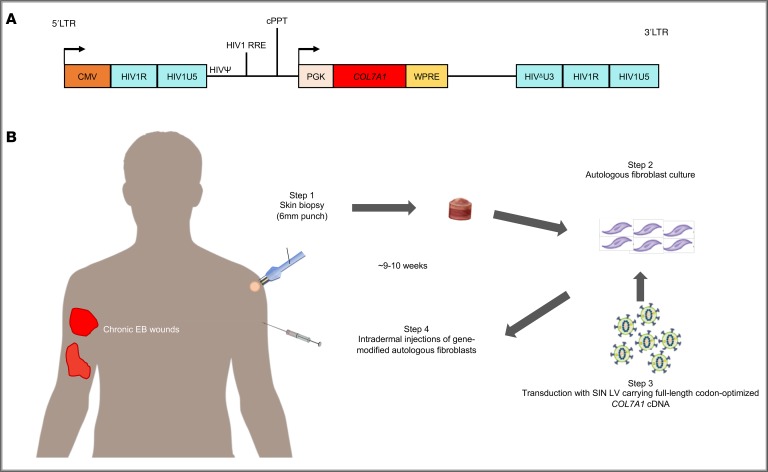
Figure 1. Illustration of Good Manufacturing Practice–compliant production of COL7A1-supplemented autologous fibroblasts for patients with recessive dystrophic epidermolysis bullosa in the present study (also referred to as the LENTICOL-F Trial).
(A) Configuration of a third-generation, 4-plasmid-system, self-inactivating lentiviral vector with deleted U3 region of the 3′ LTR, carrying full-length codon-optimized COL7A1 cDNA (SIN LV–coCOL7A1). An internal PGK promoter, mutated Woodchuck hepatitis virus posttranscriptional regulatory element (WPRE), and HIV central polypurine tract (cPPT) are shown. (B) Steps involved in the GMP manufacture of ex vivo gene-modified autologous fibroblasts for intradermal injections in a subject with recessive dystrophic epidermolysis bullosa (RDEB). It took approximately 9–10 weeks from the time of obtaining the skin biopsy sample to the time of treatment.
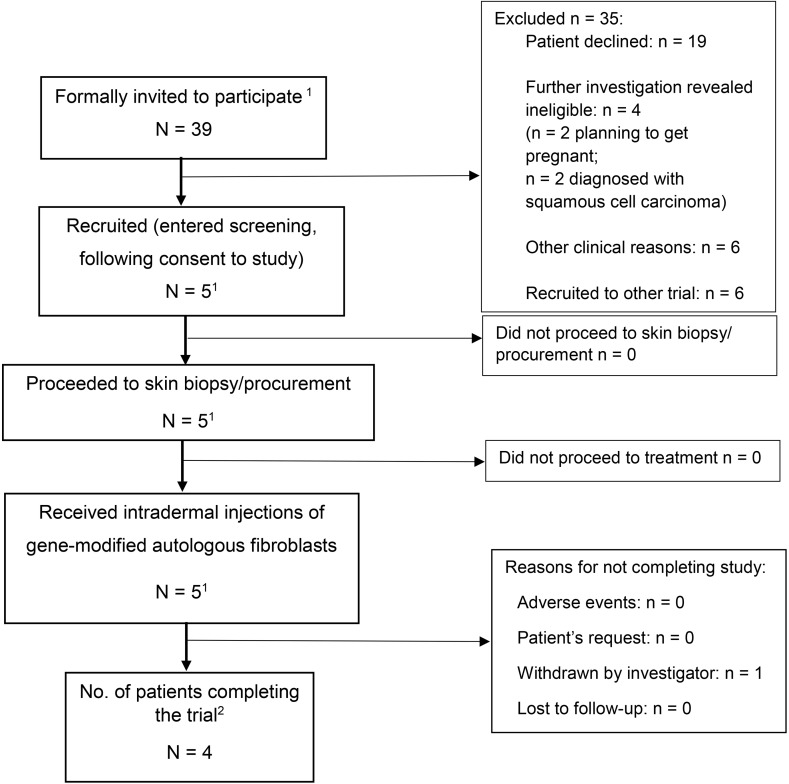
Figure 2. CONSORT trial flowchart.
1Includes subject 1 twice, as he was withdrawn and reenrolled as subject 2 due to failure of delivery of the investigational medicinal product (IMP) — i.e., gene-modified autologous fibroblasts — as per consensus from the trial steering committee members and ethics approval. 2Trial completion includes all subjects followed up for 12 months after administration of gene-modified autologous fibroblasts.
Results
Patients.
Thirty-nine RDEB adults with confirmed biallelic COL7A1 mutations by Sanger sequencing were invited to participate in the study, of which 35 patients were excluded due to reasons stated in the trial flowchart (Figure 2). The trial protocol synopsis is summarized in Supplemental Table 1 and the study timeline in Supplemental Figure 1 (supplemental material available online with this article; https://doi.org/10.1172/jci.insight.126243DS1). A total of 4 RDEB adults were subsequently enrolled to the clinical trial after providing informed consent, having met the eligibility criteria. The first patient was withdrawn 3 months after treatment and reenrolled as subject 2 due to failed delivery of gene-modified fibroblasts at first treatment. This was based on the consensus decision by the trial steering committee and approval by the gene therapy advisory committee. Thus, 4 adult patients constituted the 5 subjects for the trial. The first patient was recruited in September 2015, and the final visit of the last patient was completed in March 2018.
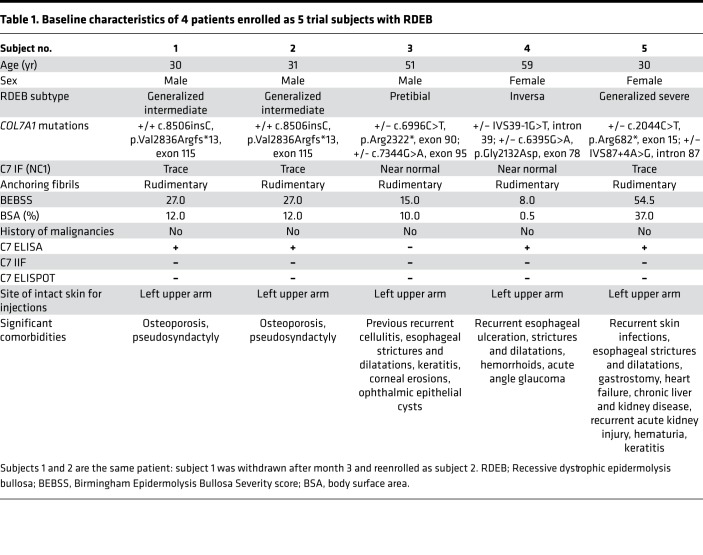
Subjects 2–5 completed all 7 follow-up visits over 12 months for the evaluation of primary and secondary end points. The primary safety endpoints were assessed for subjects 1–5, whereas the secondary efficacy endpoints were assessed for subjects 2–5 only in light of failed cell delivery in subject 1. Baseline characteristics for all 5 subjects with RDEB are summarized in Table 1; there were 2 males (one of whom was enrolled twice as subjects 1 and 2) and 2 females, with a mean age of 40.2 years (range, 30–59 years), and the patients had generalized severe, generalized intermediate, inversa, and pretibial subtypes of RDEB, with baseline Birmingham Epidermolysis Bullosa Severity scores (BEBSS) of 8–54.5 and affected body surface areas of ~0.5%–37%. Among all the subjects, subject 5 had the most severe phenotype, with the generalized severe form of RDEB. There was a wide range of comorbidities for all subjects, including esophageal and urethral strictures, pseudosyndactyly, chronic kidney and liver disease, recurrent skin infections, and left ventricular cardiac failure. All 5 subjects had intact nonblistered skin over the left upper arm; thus, intradermal injections were administered over this same site in all individuals to prevent anatomical variations in C7 expression and AF numbers (Table 1).
Table 1. Baseline characteristics of 4 patients enrolled as 5 trial subjects with RDEB.
In terms of genotype-phenotype correlations (Table 1), subject 1/2 had a homozygous single nucleotide insertion, c.8506insC (p.Val2836Argfs*13), in exon 115, leading to a frameshift and downstream PTC, resulting in truncated C7 expression. Thus, only trace staining of C7 was detected by immunofluorescence microscopy (IF) using antibodies against the NC1 domain in this subject. Subject 5 also demonstrated trace C7 expression using anti–C7 NC1 antibodies on IF in her baseline skin biopsy, as she was compound heterozygous for a nonsense mutation, c.2044C>T (p.Arg682*), in exon 15 and a donor splice site mutation, IVS87+4A>G, in COL7A1. This particular nonsense mutation occurs within the NC1 domain of C7 that leads to expression of a highly truncated C7 as well as low levels of mRNA from this allele because of nonsense-mediated RNA decay. On the other allele, the donor splice site mutation occurs at the +4 position, for which previous analysis has shown generation of a leaky splice site with some endogenous synthesis of full-length C7 or in-frame skipping of exon 87 that contains 69 bp leading to a slightly truncated polypeptide (23-amino-acid truncation) within the triple helix domain of the C7 protein (24, 25). In contrast, subjects 3 and 4 had bright C7 staining at baseline. Subject 3 was compound heterozygous for a nonsense mutation, c.6996C>T (p.Arg2322*), in exon 90 and a splice site mutation, c.7344G>A, in exon 95 (last nucleotide of the exon). Subject 4 was compound heterozygous for an acceptor splice site mutation, IVS39-1G>T, and a glycine substitution, c.6395G>A (p.Gly2132Asp), in exon 78. These combinations of mutations, all of which occur in the triple helical region of C7, allow for expression of some truncated or structurally altered proteins when assessed using antibody staining with LH7.2 against the NC1 domain of C7.
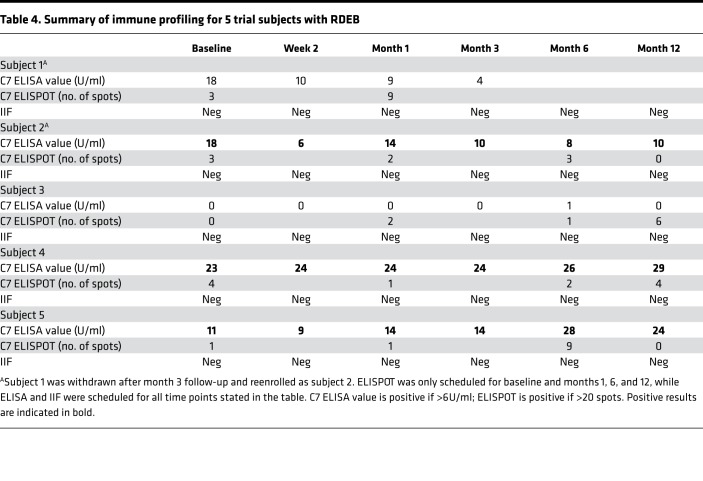
Transmission electron microscopy (TEM) analysis of baseline skin biopsies from all subjects demonstrated that none of the study participants had centrosymmetric, fan-shaped mature AFs but instead had wisp-like structures of rudimentary AFs. In terms of autoimmune reactions against recombinant C7, all subjects except subject 4 had a positive C7 ELISA (value: 11–23 U/ml), but all had negative indirect immunofluorescence (IIF) and ELISPOT at baseline. These observations suggest that all but subject 4 had circulating autoantibodies against C7 (for both NC1 and NC2 domain epitopes), but none bound to the basement membrane of salt-split skin to potentially result in the pathogenic acquired form of EB known as EB acquisita (EBA) (26).
Restoration of C7 expression in Good Manufacturing Practice–compliant SIN LV–COL7A1–transduced primary RDEB fibroblasts.
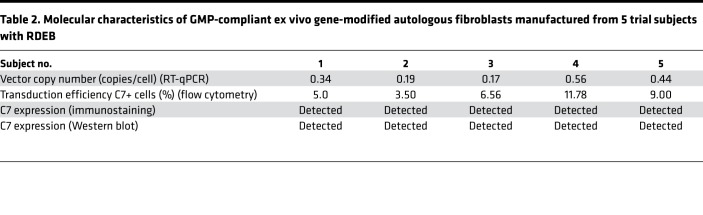
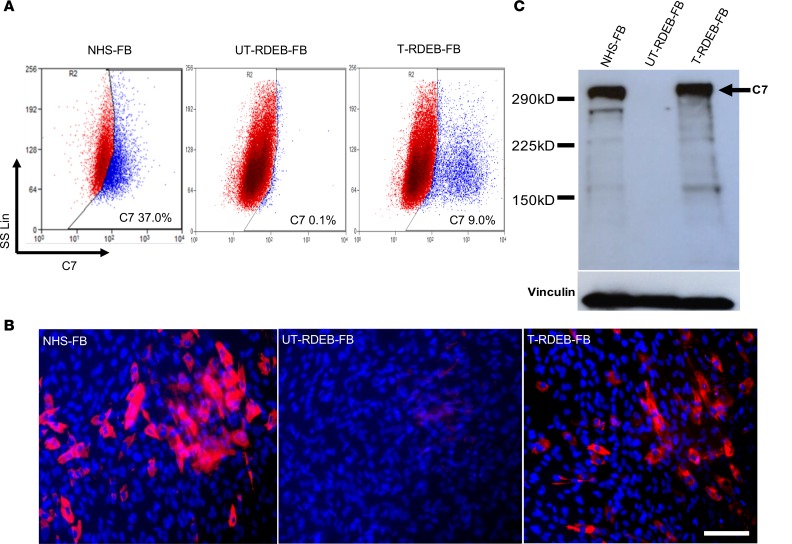
Autologous primary RDEB fibroblasts were transduced with clinical grade third-generation SIN LV–PGK–COL7A1 virus using a single round of exposure at an MOI of 5 in accordance with Good Manufacturing Practice (GMP) standards. Five gene-modified autologous fibroblast products were manufactured (summarized in Table 2) and had a mean vector copy number (VCN) of 0.34 copies/cell (range, 0.17–0.56 copies/cell) and a mean C7 expression on flow cytometry analysis of 7.2% (range, 3.5%–11.8%) (Figure 3A). In situ cytofluorescence staining against the C7 NC1 domain detected protein expression in transduced RDEB fibroblasts (Figure 3B), whereas C7 was absent in untransduced RDEB fibroblasts. These results were further confirmed by Western blot analysis using a purified C7 antibody. Transduced RDEB fibroblasts at ~5 weeks after transduction revealed expression of a ~290-kDa protein band corresponding to full-length C7 (Figure 3C).
Table 2. Molecular characteristics of GMP-compliant ex vivo gene-modified autologous fibroblasts manufactured from 5 trial subjects with RDEB.
Figure 3. Molecular characteristics of GMP-manufactured gene-modified autologous fibroblasts for subject 5.
Primary fibroblasts from 5 subjects. Four 4 patients in the trial were transduced with LV-PGK-coCOL7A1 at an MOI of 5, and the expression of C7 in transduced RDEB fibroblasts was assessed 4 weeks after transduction. (A) In subject 5, 9.0% of the transduced fibroblasts expressed C7 compared with untransduced RDEB fibroblasts as assessed by flow cytometry. (B) In situ cytoimmunofluorescence staining confirmed C7 (red: C7 and blue: nuclei) expression in both LV-PGK-coCOL7A1–transduced fibroblasts and normal human skin fibroblasts but not in untransduced RDEB fibroblasts. Scale bar: 50 μm. (C) C7 expression was further confirmed by immunoblotting, and results showed 290-kDa full-length C7 expression in transduced RDEB fibroblasts and normal human skin cells but not in untransduced fibroblasts. Vinculin represents internal loading control. NHS-FB, normal human skin fibroblasts; UT-RDEB-FB, untransduced RDEB fibroblasts; T-RDEB-FB, RDEB fibroblasts transduced with LV-PGK-coCOL7A1.
Intradermal injections of COL7A1-modified autologous fibroblasts.
Each subject received 3 intradermal injections (1.14 × 106 to 1.2 × 106 cells/cm2 of intact skin) of gene-modified autologous fibroblasts on day 0, with each injection administered over a 1-cm2 area of skin, with point tattooing to mark the injection sites (Supplemental Figure 2). Patients were monitored for vital signs at 15 minutes and 30 minutes after the injections to assess any immediate reactions to gene-modified fibroblasts, but no serious adverse reactions (SARs) were observed after treatment. Both primary safety and secondary efficacy endpoints were assessed at various time points on the injected skin over 12 months (Supplemental Figure 1). All patients received standard care for symptomatic treatments as required during this trial, such as esophageal dilatation.
Safety.
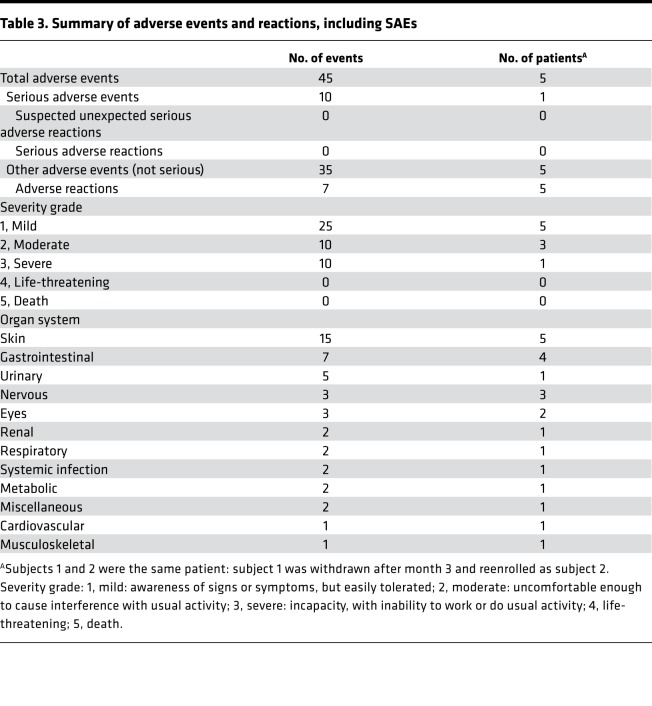
All 5 subjects (4 patients) tolerated the injections without SARs but developed 7 mild (grade 1) adverse reactions (ARs) related to the administrative procedure but not the gene-modified fibroblasts themselves over the 12 months. These included injection site erythema (n = 4), pain (n = 1) and bruising (n = 1), and itch over the skin biopsy site (n = 1), all of which resolved spontaneously without treatment. A total of 45 adverse events (AEs) in 5 subjects (Table 3) were graded as mild (grade 1; n = 25), moderate (grade 2; n = 10), and severe (grade 3; n = 10). The most common AEs were EB-related skin conditions (n = 15), followed by gastrointestinal conditions (n = 7) and urinary conditions (n = 5). Subject 5 experienced 10 serious AEs (SAEs) over 3 hospital admissions secondary to preexisting comorbidities rather than the treatment or injection itself. There were no life-threatening (grade 4) events or deaths (grade 5). Vital signs, physical examination, clinical photographs of the injection sites, and blood analyses, including full blood count, urea and electrolytes, liver function tests, and C-reactive protein, were also performed at each visit, both before and after the treatment, except at week 1 and month 9 (Supplemental Figure 1). There were no clinically important changes in any of these measures compared with respective individual baselines to indicate any safety concerns throughout the duration of the trial. Over the 12 months of monitoring, no subject developed malignancy after injection; no tumorous cells were evident by light microscopy (data not shown). In terms of immunogenicity, there was no evidence of pathogenic autoimmune reactions against recombinant C7 (Table 4). There were no clinically significant changes in anti-C7 IgG values on ELISA, except in subject 5, in whom the value doubled at month 6 compared with month 3, although the baseline was also positive (14 U/ml at month 3 to 28 U/ml at month 6; baseline, 11 U/ml) but began to decrease at month 12 (24 U/ml). This observation coincided with an increase in C7 ELISPOT from 1 spot at baseline and month 1 to 9 spots at month 6, but with a rapid decrease to 0 spots by month 12, albeit all results being negative at all time points assessed. However, none of the circulating autoantibodies against C7 bound to the basement membrane, as evidenced by negative IIF in all 4 adults (subjects 2–5) at baseline, 2 weeks, 4 weeks, 3 months, 6 months, and 12 months after gene therapy. Therefore, there was no evidence of pathogenic EBA in these patients secondary to gene-modified fibroblasts. The C7 ELISPOT also remained negative for all 4 adults (subjects 2–5) at baseline, 4 weeks, 6 months, and 12 months after gene therapy, indicating lack of cytotoxic T cell response against newly synthesized C7 in those who had increased C7 expression. Subject 1 also had negative IIF and ELISPOT at all time points up to month 3, after which he was withdrawn from the trial due to treatment delivery failure, as described above.
Table 3. Summary of adverse events and reactions, including SAEs.
Table 4. Summary of immune profiling for 5 trial subjects with RDEB.
Efficacy: C7 expression following intradermal injections of gene-modified autologous fibroblasts in RDEB patients.
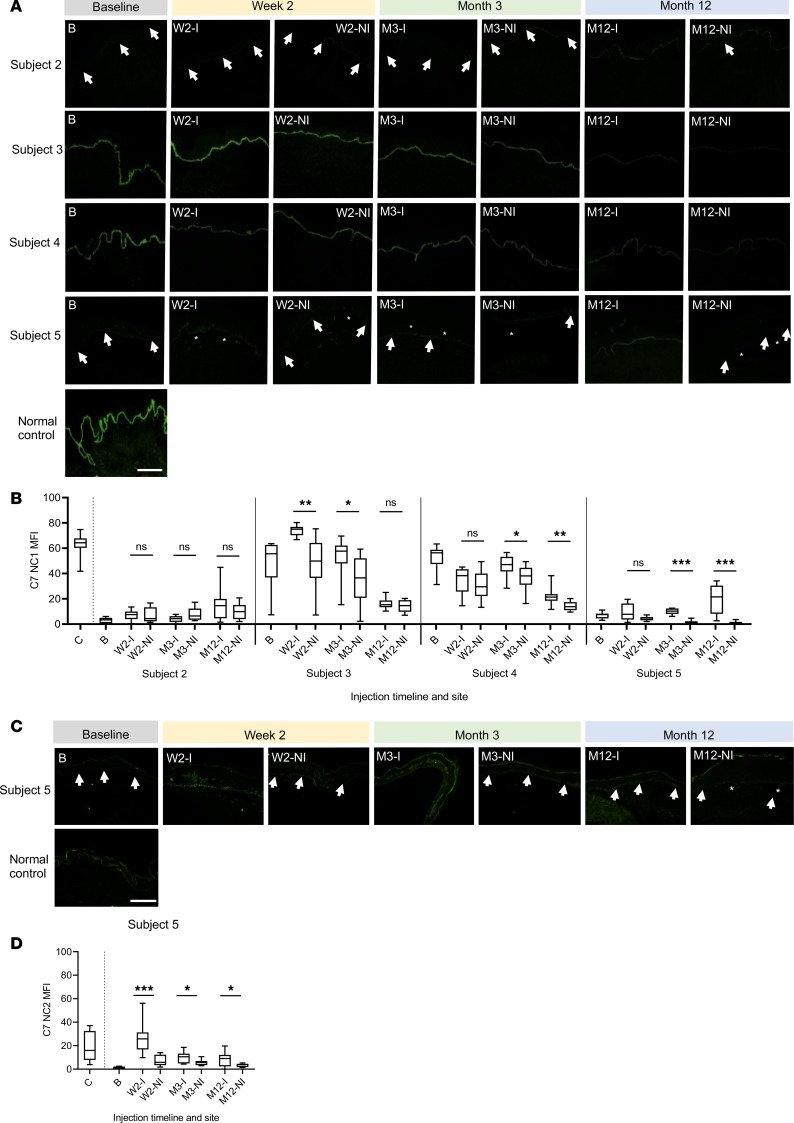
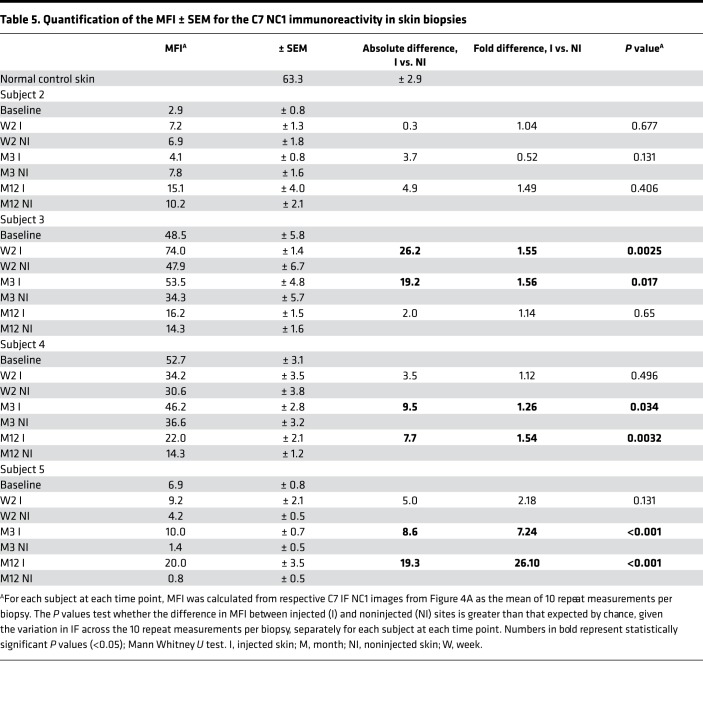
MFI of C7 NC1 staining was calculated from 10 measurements taken at regular intervals along the DEJ from the C7 IF images of both injected skin and noninjected skin samples, at all 3 time points, for subjects 2–5 (Figure 4, A and B, and Table 5) in an assessor-blinded manner using ImageJ (NIH) (24). The changes in the C7 MFI are illustrated in Figure 4B. Overall, there was an increase in C7 expression in 11 out of 12 skin biopsies taken from the injected sites in 4 subjects compared with biopsies from noninjected sites at 2 weeks, 3 months, and 12 months after injections (subject 1 was excluded from analysis due to failed delivery of gene-modified fibroblasts). A statistically significant increase in C7 MFI was detected in the injected skin when compared with noninjected skin in 6 of 12 comparisons for 3 of 4 subjects analyzed, ranging from a 1.26-fold increase at 3 months in subject 4 (46.2 vs. 36.6, P = 0.034) to a 26.10-fold increase at 12 months in subject 5 (20.0 vs. 0.8, P < 0.001). (Figure 4, A and B, and Table 5). There was a nonsignificant increase in C7 MFI in the injected skin compared with noninjected skin in 5 of 12 comparisons in all 4 subjects analyzed. There was also 1 nonsignificant decrease in C7 MFI by 0.52-fold in the injected skin compared with noninjected skin in subject 2 at 3 months (4.1 vs. 7.8, P = 0.13).
Figure 4. Type VII collagen (C7) expression in subjects 2–5 at baseline and in injected and noninjected skin at 2 weeks, 3 months, and 12 months.
(A) IF staining with a monoclonal LH7.2 antibody to C7 (targeting NC1 domain) on skin sections taken at specified time points from both injected (I) and noninjected (NI) sites alongside respective baseline samples. Trace C7 staining is visible in subjects 2 and 5, while brighter staining is seen in subjects 3 and 4. White arrows indicate the trace staining of C7 IF; asterisks indicate dermal-epidermal separation (B) The MFI of C7 NC1 expression along the DEJ in the injected and noninjected skin demonstrated in A for subjects 2–5 was further quantified for each sample in an assessor-blinded fashion using ImageJ, as adapted from a previous study (27). An average MFI value was calculated from 10 measurements taken at regular intervals using 8 × 8 pixels every 100 pixels along the DEJ. The C7 NCI MFI graphs in B correspond to the C7 IF images in A. (C) IF staining to demonstrate full-length C7 expression in subject 5 using a polyclonal antibody targeting the NC2 domain of C7 protein (ab198899; Abcam) after optimization. (D) The C7 NC2 MFI was then calculated using ImageJ as for the C7 NC1 MFI as described in B in an assessor-blinded fashion. The box-and-whisker plots in B and C represent the 10 repeat measurements of C7 MFI taken along the DEJ. *P < 0.05, **P < 0.005, ***P < 0.001. Error bars represent mean ± SEM. White arrows indicate C7 expression at the DEJ. B, baseline; C, control (normal skin); I, injected skin; M, month; NI, non-injected skin; W, week. Scale bars: 100 μm.
Table 5. Quantification of the MFI ± SEM for the C7 NC1 immunoreactivity in skin biopsies.
Expression of full-length C7 (i.e., C7 NC2) in subject 5 following intradermal injections of gene-modified autologous fibroblasts.
In order to analyze whether the significantly increased C7 expression in subject 5 was full length, we performed C7 IF using a commercially available polyclonal antibody targeting the NC2 domain of C7 protein (ab198899; Abcam). The C7 NC2 MFI was then calculated using ImageJ as for the C7 NC1 MFI, as described above in an assessor-blinded manner (24). Subject 5 was chosen for 2 reasons: first, she had a compound heterozygous nonsense/splice site combination of mutations expressing endogenous WT full-length C7 as well as mutant truncated C7; second, she reported her observation that the biopsy site from the injected skin healed much faster than the biopsy site from noninjected skin. In the injected skin compared with noninjected skin, there was a 3.6-fold increase in full-length C7 expression at 2 weeks (26.6 vs. 7.4, P < 0.001), 1.8-fold increase at 3 months (10.0 vs. 5.6, P = 0.041), and 2.5-fold increase at 12 months (8.3 vs. 3.3, P = 0.034) after treatment (Figure 4, C and D).
No AFs were detected in the injected skin despite increased C7 expression after treatment.
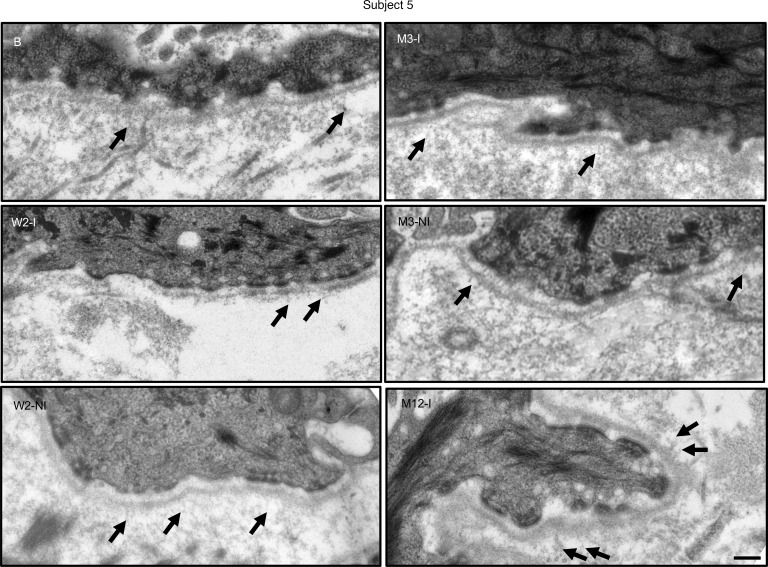
TEM analysis of the basement membrane zone in injected skin of subjects 2–5 did not reveal any mature fan-shaped AFs in any of the skin biopsies from injected sites at week 2, month 3, or month 12, although wisp-like structures (possible immature/rudimentary AFs) similar to those seen at baseline were present (Figure 5 and Supplemental Figure 3).
Figure 5. Transmission electron microscopy failed to demonstrate mature AFs along the dermal-epidermal junction of subject 5.
Thin sections of upper left arm control baseline (B), injected (I), and noninjected (NI) skin from subject 5 were obtained at 2 weeks, 3 months, and 12 months after treatment. No mature fan-shaped AFs were observed on any micrographs, but occasional thin wisp-like rudimentary AFs (black arrows) were visible at all time points. The epidermis was detached upon skin biopsy of the noninjected skin at month 12, and thus we were unable to obtain a meaningful intact dermal-epidermal junction transmission electron microscopy images. B, baseline; I, injected skin; M, month; NI, noninjected skin; W, week. Scale bar: 0.25 μm.
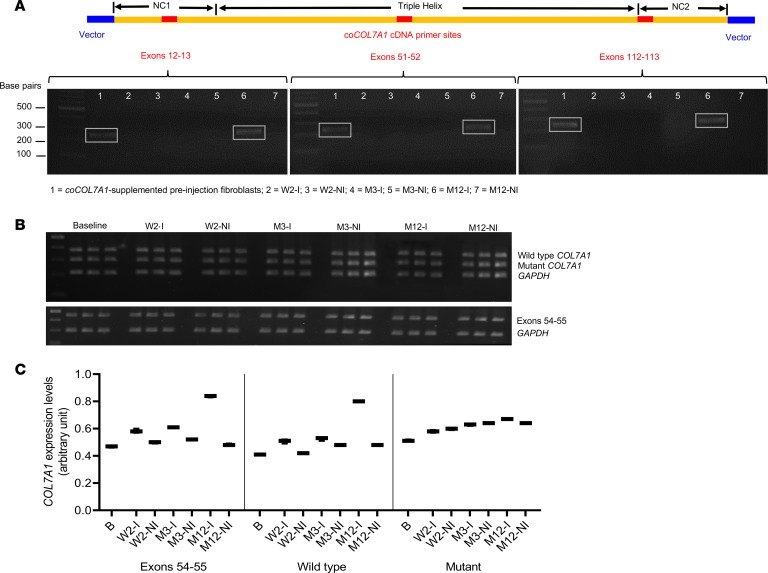
Detection of COL7A1 transgene (codon-optimized COL7A1 cDNA) in the injected skin at 12 months after gene therapy in subject 5.
As previously stated, subject 5 harbored a compound heterozygous nonsense/splice site combination of mutations in COL7A1 expressing both truncated and WT full-length C7. Therefore, it was pertinent to evaluate whether the increase in full-length C7 expression in the injected skin of subject 5 at week 2, month 3, and month 12 were accounted for by the presence of transgene in vivo. To do this, we performed PCR amplifications using 3 sets of primers targeting the NC1, triple helix, and NC2 regions unique to the codon-optimized COL7A1 transgene sequence (coCOL7A1 cDNA) (exon 12–13, exon 51–52, and exon 112–113, respectively) that is different from the endogenous WT COL7A1. This analysis demonstrated 3 clear bands at month 12 in the injected skin and transduced autologous fibroblasts before injection but not for the noninjected skin or baseline skin; there was a band at 247 bp corresponding to the exon 12–13 fragment of coCOL7A1, at 231 bp corresponding to the exon 51–52 fragment, and 244 bp corresponding to the exon 112–113 fragment of coCOL7A1 (Figure 6, A–C). This finding suggests that the increased full-length C7 expression was at least in part due to gene-modified autologous fibroblasts, which were detectable at month 12 after treatment in subject 5. The same PCR amplifications were performed for subjects 2, 3, and 4 for skin samples taken at week 2, month 3, and month 12 after treatment, but no coCOL7A1 was detected (data not shown). Conversely, when the VCN was measured using quantitative PCR (qPCR) with primers specific to the packaging signal (psi) sequences of the LV, none was detected in any of the skin biopsies of subjects 2–5 (data not shown).
Figure 6. PCR and semiquantitative PCR demonstrating the expression of COL7A1 transgene and endogenous mutant and WT cDNA respectively in the injected skin of subject 5.
(A) PCR amplifications using 3 sets of primers targeting NC1, triple helix, and NC2 regions unique to codon-optimized COL7A1 transgene sequence (coCOL7A1 cDNA) (exon 12–13, exon 51–52, and exon 112–113, respectively) that are different from the endogenous WT COL7A1 cDNA sequence. This demonstrated clear bands (in boxes) at month 12 (lane 6) in the injected skin and transduced autologous fibroblasts before injection but not the noninjected skin or baseline skin; there was a band at 247 bp corresponding to the exon 12–13 fragment of coCOL7A1, at 231 bp corresponding to the exon 51–52 fragment, and 244 bp corresponding to the exon 112–113 fragment of coCOL7A1. Similar clear bands (in boxes) were also seen in the gene-modified fibroblasts (i.e., coCOL7A1-supplemented) before injections (lane 1), which represent the positive control. (B) Multiplex PCR amplifications of endogenous COL7A1 cDNA reversed transcribed from the total RNA extracted from the injected and noninjected skin using specific primers to detect mutant exon 87–skipped (without exon 87) sequence and WT (with exon 87) sequence of COL7A1 with GAPDH as internal control. Each band on the PCR gel image was used for quantification with GelQuant.NET. (C) Semiquantitative PCR demonstrated a relative increase in expression of both endogenous WT and mutant COL7A1 in the injected skin of subject 5 (WT at week 2, month 3, and month 12 after gene therapy, and mutant at month 12 after gene therapy), after normalization with the housekeeping gene GAPDH. The box-and-whisker plot represents triplicate measurements of respective COL7A1 expression. B, baseline; I, injected skin; M, month; NI, noninjected skin; W, week.
Discussion
To date, only 3 human studies of ex vivo gene therapy in the form of genetically corrected keratinocyte sheets in EB are have been published to our knowledge: 2 reports on junctional EB (JEB) (27, 28), but only 1 study with 4 adults on RDEB (29). The RDEB trial demonstrated acceptable safety with variable efficacy that declined over 1 year (29). In contrast, long-term therapeutic efficacy was demonstrated in a pediatric patient with JEB after successfully targeting the holoclone keratinocyte stem cell population with a LAMB3 transgene (28). Practically, however, keratinocytes grafted onto wounds are inherently fragile and may be prone to loss through trauma or infection, especially in RDEB, as skin wounds in these patients are typically very inflamed and often colonized or infected with numerous bacteria or yeasts. By contrast, intradermal injections of fibroblasts into intact, unwounded RDEB skin offers a “closed” system, one that is not influenced by external organisms or other factors that may damage or destroy fragile grafts. Additionally, the intradermal injection system allows a more practical and feasible treatment of gene therapy for RDEB in the clinical setting, where the treatment can be delivered in an outpatient setting by the clinician without the need for invasive surgery, possibly under general anesthesia.
To our knowledge, the results from the trial we present here is the first human study demonstrating safety and potential efficacy of SIN LV–mediated gene-modified autologous fibroblasts for RDEB that follows on from encouraging preclinical in vivo data (22). All 4 adults tolerated 3 intradermal injections of SIN LV–COL7A1–modified fibroblasts without SARs, with only mild local injection procedure-related side effects that lasted for a few hours and without requiring treatment.
For RDEB subjects, especially those with null mutations, ex vivo gene therapy poses a theoretical risk of immune response against the newly synthesized full-length C7 (30, 31). The C7 ELISA, ELISPOT, and IIF data from this study suggest that none of the subjects developed a pathogenic immune response, EBA, after treatment. Subject 5’s ELISA value and ELISPOT count transiently raised at month 6 compared with previous values at month 3 and baseline but remained below the threshold of positivity. However, when subject 5’s sera were placed on salt-split skin, there was no evidence of circulating anti-C7 antibodies binding to the basement membrane, indicating a lack of pathogenicity against newly synthesized C7. This observation is in agreement with previous studies using gentamicin as a PTC-readthrough drug, expressing full-length C7 (14), and retrovirus-mediated ex vivo transduced keratinocyte sheets (29). Nevertheless, further evaluation of longer-term safety of ex vivo gene therapy is warranted in trials with a larger sample size involving patients with a variety of genotypes, including those with null mutations and any dose-dependent antibody response against recombinant C7.
Regarding efficacy, there was a significant increase in C7 expression at the DEJ in 6 of 12 injected skin sites compared with noninjected sites in subjects 2–5 (Figure 4 and Table 5), with 2 of these 4 subjects showing a significant increase at 12 months, after a single intradermal injection with ~1 × 106 cells/cm2 of skin. However, the level of increase in C7 expression was variable, and no associated mature AFs were visualized, nor were VCNs detected in the injected skin by qPCR. One possible explanation for the failure to develop mature AFs after gene therapy may lie within the posttranslational steps of procollagens. Procollagens are catalyzed by lysyl hydroxylases (LHs), including LH3 that is present in the subepidermal extracellular space and colocalized with C7 at the DEJ (32). We do not know the state of LH3 expression in these trial subjects, and the treatment in question is not designed to optimize LH3 activity. Another probable explanation is that other potential paracrine factors such as extracellular vesicles may be suboptimal, as the target cells are fibroblasts and not MSCs, which are well known to release these extracellular vesicles. The latter help transport C7 within the extracellular space (13), where they assemble into a centrosymmetric dimer arrangement with carboxyl terminal tail-to-tail organizations, which then laterally aggregate to form AFs (33). Studies of skin regenerated from cultured epithelial autografts on full-thickness burns illustrated that AFs can reform in a punctate fashion after 6 days, but complete maturation requires more than a year (34). This may explain the lack of mature AFs up to 12 months despite significant C7 restoration in the area of skin injected with gene-modified fibroblasts. Another possible explanation is that the amount of increase in C7 protein expression was still subtherapeutic for the AF formation, owing to limited cell numbers injected (~1 × 106 cells/cm2).
During the trial, subject 5 voluntarily reported her own observation that the biopsy site from the injected skin healed much faster than the biopsy site from noninjected skin. This participant’s observation was consistent with the fact that there was a significant increase in full-length C7 expression in her treated skin compared with untreated skin at all time points assessed (i.e., week 2, month 3, and month 12) (Figure 4). Interestingly, in subject 5, the largest increase in anti–NC1 C7 MFI was detected at month 12 (Figure 4, A and B), while the largest increase was detected at week 2 for anti-NC2 C7 MFI (Figure 4, C and D). This indicates that some of the C7 IF detected by anti-NC1 antibody was not full-length, and the increase in full-length C7 MFI was accounted for not just by the COL7A1-modified fibroblasts, but potentially also by subject 5’s endogenously expressed mutant and WT COL7A1. Indeed, using a combination of PCR and semiquantitative multiplex PCR techniques, we were able to illustrate both the expression of the coCOL7A1 transgene at month 12 and a relatively small increase in expression of endogenous WT COL7A1 at week 2, month 3, and month 12, and mutant (exon 87–skipped) COL7A1 at month 12 after gene therapy, as analyzed from total RNA extracted from the injected skin of subject 5 compared with her noninjected skin (Figure 6). The PCR amplifications using primers specifically targeting coCOL7A1 cDNA demonstrated 3 clear bands at 247 bp corresponding to the exon 12–13 fragment of coCOL7A1 (NC1 domain of the C7 protein), at 231 bp corresponding to the exon 51–52 fragment (triple helix domain of C7), and 244 bp corresponding to the exon 112–113 fragment of coCOL7A1 (NC2 domain of C7) (Figure 6A). These findings suggest that the increased expression of C7 in subject 5 was at least in part due to coCOL7A1 transgene expression in addition to endogenous expression of predominantly WT COL7A1 (Figure 6, B and C). Production of mostly WT C7 from subject 5’s endogenous COL7A1 is expected because of the leaky donor splice site mutation at the +4 position (24). These findings support the rationale for the use of autologous cells for a longer-lasting therapeutic effect, as opposed to allogenic cells, which were rejected within 2 weeks after intradermal injections, with upregulation of C7 secondary to complex autocrine effects involving heparin-binding epidermal growth factor–like growth factor (HB-EGF) up to 9 months (25). The efficacy of gene-modified fibroblasts may be partly mediated through local paracrine effects triggered by the intradermal injections and saline itself (24, 25), but more importantly by the secretion of recombinant C7 produced in situ by a subpopulation of transduced cells (Figure 6). Similarly, in subjects 3 and 4, there was a significant increase in C7 NC1 expression at 2 weeks and 3 months, and 3 months and 12 months, respectively. This may be partly attributed to a possible increase in endogenous expression of truncated proteins compared with baseline via an HB-EGF–mediated autocrine effect. This is in contrast to subject 2, who did not have any significant increase in C7 expression after injections.
Another interesting finding is that although the conventional PCR technique allowed us to detect the coCOL7A1 transgene in the injected skin (and not in the noninjected skin) of subject 5, it was only present at month 12 and not at week 2 or month 3 (Figure 6A). Moreover, the qPCR analysis of VCN failed to detect any proviral copy in the injected skin of subject 5 at any of the time points. These findings are curious and perhaps could be explained by the variability in numbers of engrafted gene-modified fibroblasts within the injected areas. Another explanation could be the relative difference in sensitivity of assays used, i.e., conventional PCR versus qPCR. Indeed, it has been reported that conventional PCR may be more sensitive than qPCR in some cases (35, 36). While the transgene may account for the increase in C7 MFI (both NC1 and NC2) in the injected skin (Figure 4) of subject 5 at month 12, PCR failed to detect the presence of coCOL7A1 transgene for subject 5 at week 2 and month 3. This may reflect the time it took for the autologous fibroblasts that remained viable after the intradermal injections to undergo mitosis to reach a large enough cell population in vivo for detection by conventional PCR.
Nevertheless, only modest efficacy was observed in other subjects. Subject 2, carrying PTC mutations, did not express any significantly increased C7, whereas subjects 3 and 4 had increased C7 synthesis at 2 of 3 time points analyzed, with limited and transient efficacy. Although fibroblasts are considered to be more robust, exhibit less differentiation and growth arrest (37), and have the ability to contribute more C7 expression at the DEJ compared with keratinocytes (20), they tend to senesce rapidly in vivo (37). This nature of fibroblasts, together with low cell numbers delivered intradermally, might account for the limited efficacy. Interestingly, a recent study by Hirsch and colleagues (28) elegantly demonstrated that epidermal stem cell holoclones (~1,888 holoclones/cm2) were responsible for the therapeutic longevity of transgenic keratinocyte grafts in a pediatric patient with JEB. Another possible explanation for the limited efficacy is the epigenetic modification of the promoter through DNA methylation, which can lead to transgene silencing over time, especially with oncoretroviral systems (33, 37); for review, see ref. 38. Indeed, the PGK promoter used to drive expression of coCOL7A1 in the LV used in this trial has been shown to become silenced over time within an LV context following delivery to hematopoietic stem cells (39). The safety of the LV-PGK-COL7A vector was optimized through the use of a human PGK internal promoter and codon-optimized COL7A1 transgene with eliminated cryptic splice sites. However, the modest vector titer and resultant transduction efficiency owing to the large cargo size of 9.2 kb might have further accounted for the limited efficacy.
Limitations of this study included a small sample size; safety was assessed in all subjects (subject 1 up to month 3 and subjects 2–5 up to month 12), whereas efficacy endpoints were evaluated for subjects 2–5 only. This is due to subject 1 being withdrawn from the study at 3 months after treatment based on the consensus decision of the trial steering committee as a result of failed delivery of gene-modified cells, which appeared as turbid clumped cells in clear 0.9% saline solution at time of injections. The corrective action plan (gently tapping the syringe to evenly resuspend the clumped cells prior to injections) was immediately executed, with successful delivery of gene-modified cells in all subsequent subjects. Owing to the limited availability of local GMP facilities and the lengthy transduction process (duration of ~9–10 weeks), the minimal essential number of cells that would provide clinically meaningful safety outcomes was extrapolated from previous reports (40), with the cell dose determined to be ~1 × 106 cells/cm2.
This was primarily a safety trial; therefore, the gene-modified cells were injected in the intact, nonblistered skin only to enable clear visualization of any local reactions at the site of injection, such as erythema. The study was also not sufficiently powered or blinded for the assessment of secondary efficacy endpoints, which included primarily molecular assessments such as C7 expression and AF formation; these did not include clinical measures such as wound healing or the strength of dermal-epidermal adherence via quantitative measures, e.g., suction blister time, due to limited surface areas of treated skin. Additionally, the full extent of the potential efficacy of gene-modified fibroblasts, if any, could not have been evaluated with the limited number of cells delivered (~1 × 106/cm2). Neither replication-competent lentivirus (RCL) nor integration site analyses were performed, since there were no SARs or development of cutaneous squamous cell carcinoma in any of the 15 injected skin sites. It should also be noted that the statistical tests performed were exploratory, and we cannot rule out that some of the significant P values may have been the result of multiple testing; however, the nonsignificant results showed a similar trend toward increased C7 levels in the injected sites.
In summary, despite these limitations, we have demonstrated proof-of-concept delivery of gene-modified autologous fibroblasts via intradermal injections in 5 adults with RDEB, alongside safety and molecular efficacy with increased C7 expression in 6 of 12 injected skin sites. To the best of our knowledge, this is also the first study to demonstrate the presence of COL7A1 transgene in the injected skin at 12 months after treatment in 1 patient with an associated increase in full-length C7 expression at the DEJ. These data provide the foundation to develop controlled phase II trials with larger cell numbers in both adults and children with RDEB for further evaluation of the potential long-term safety and efficacy of gene-modified autologous fibroblasts.
Methods
Further information can be found in Supplemental Methods, available online with this article.
Study design and participants.
This was an open-label, single-center phase I study that aimed to assess the safety and early efficacy of ex vivo lentiviral gene-modified autologous fibroblasts in adults with RDEB. There were 2 screening visits, 1 treatment visit, and 7 follow-up visits, as illustrated in Supplemental Figure 1. The trial was conducted at Guy’s and St Thomas’ (GSTT) NHS Foundations Trust Clinical Research Facility (CRF) and outpatient clinics, London, United Kingdom. The COL7A1-modified autologous fibroblasts were manufactured at the GMP-compliant Gene and Cell Therapy Facility at Great Ormond Street Hospital (GOSH), London. Patients aged 17 years or older with a clinical diagnosis of RDEB were identified from the EB patient database at GSTT. Patients with confirmed biallelic mutations in COL7A1 by Sanger sequencing were then invited to give consent for screening to participate in the clinical trial. The inclusion criteria for this study were (i) a reduced number or morphologically abnormal AFs confirmed by TEM; (ii) the presence of at least 5 × 3 cm intact skin on the trunk and/or extremities suitable for cell injections; and (iii) the ability to undergo local anesthesia. The exclusion criteria were (i) investigational medicinal products (IMPs) within the past 6 months; (ii) biopsy-proven skin malignancy; (iii) immunotherapy, including oral corticosteroids (prednisolone >1 mg/kg) for >1 week or chemotherapy within the past 60 days; (iv) known allergy to any of the constituents of the IMP; (v) positive serum antibodies to C7 confirmed by ELISA and positive IIF; (vi) positive HIV, hepatitis B/C, human T cell lymphotropic virus (HTLV), or syphilis serology; (vii) pregnancy or women of child-bearing potential who were not using an agreed means of contraception for up to 12 months after treatment. Those eligible, after providing written informed consent, donated a 6-mm punch biopsy of skin for the GMP manufacturer of ex vivo transduced autologous fibroblasts. Subject 1 was withdrawn 3 months after the injections due to suboptimal delivery of gene-modified cells based on consensus decision by the trial steering committee and subsequently reenrolled as subject 2 as approved by the gene therapy advisory committee.
Study interventions.
Each of 5 subjects with RDEB enrolled in the study received 3 intradermal injections (1.14 × 106 to 1.2 × 106 cells/cm2 of intact skin) of gene-modified autologous fibroblasts on the left upper arm on day 0 (Supplemental Figure 2). On the day of treatment, after monitoring of vital signs and blood of the trial subject, the investigator identified and agreed with the subject on a suitable site of intact skin for treatment. Corners of 3 adjacent squares, each measuring 1 cm × 1 cm,s were dotted with a surgical marker pen, after which the marked skin was gently pricked with an ink tattoo marker to create semipermanent marks to note the sites of injection for follow-up assessments. Meanwhile, at the GMP laboratory, the gene-modified fibroblasts were thawed, washed, counted, and suspended in 0.25 ml of 0.9% sodium chloride packed in a 3 × 1–ml syringe with Luer-Lok screw caps. The labeled syringes were transported with a temperature monitor from the GMP center at GOSH to the CRF at Guy’s Hospital within 4 hours of the IMP release, by a specialized courier at ambient temperature. To prevent cell clumping as in subject 1, after all checks were completed, syringes containing the gene-modified cells were gently tapped with a finger to ensure an even suspension of gene-modified cells in the saline solution prior to intradermal injections in subjects 2–5. Subjects were monitored for immediate ARs for a half an hour after treatment.
Safety assessments.
The full trial schedule including all procedures and interventions is described in Supplemental Figure 1. Briefly, assessments of adverse events, concomitant medications, physical examination, and vital signs were performed at each visit after trial enrolment. Clinical photographs of the injected skin were taken at each visit before and after injections to monitor changes in the treated skin, especially with regard to any suspicious lesions or ARs locally. A number of blood tests including full blood count, urea and electrolytes, liver function tests, C-reactive protein, and erythrocyte sedimentation rate were performed at 2 weeks, 4 weeks, 3 months, 6 months, and 12 months after treatment to assess any potential systemic reactions. Blood samples were also archived for potential future assessments of replication-competent lentivirus in the event of SARs. C7 ELISA, ELISPOT, and IIF were also measured at several time points after treatment for comparison with respective baseline values.
C7 ELISA and IIF.
Circulating anti-C7 antibodies were measured using the MESACUP kit (MBL International Corp.) according the manufacturer’s protocol. The kit measures antibodies against the NC1 and NC2 domains of C7. The signal detected at antibody values of >6 U/ml is considered positive. In IIF, patients’ sera were exposed to 4-μm sections of salt-split human skin and subsequently stained with FITC-labeled antibodies directed against human IgG (Agilent Technologies, 1:200) to determine any basement membrane–bound autoantibodies.
C7 ELISPOT.
This assay was performed as previously described (30). A clear 96-well MultiScreen filter plate (Millipore) was soaked in 1 μl of 35% ethanol (ETOH), and the wells were washed with PBS 3 times, while thoroughly shaking the plates to discard the PBS between the washes. The anti–IFN-γ capture antibody (purified mouse anti–human IFN-γ) was diluted to a final concentration of 1:50 in PBS. 50 μl of diluted capture antibody was added to each of the allocated wells, and the plate was sensitized overnight at 4°C. Adherent monocytes and CD4+ and CD8+ T lymphocytes were purified from buffy coats using 2-hour incubation in petri dishes and anti-CD4 and anti-CD8 MACS magnetic cell sorting (Miltenyi Biotec), respectively. Patient and control CD4+ and CD8+ T cells were incubated 40 hours in RPMI medium with 5% human AB serum with purified recombinant C7 (produced in-house) (10 μg/ml) or concanavalin A (3 μg/ml, Sigma-Aldrich, C5275) in ELISPOT plates coated with anti-cytokine antibodies (anti–IFN-γ, clone NIB42, or anti-IL-4, clone 8D4-8; both from BD Pharmingen), in the presence of IL-2 (50 UI/ml). Cytokine production was detected in situ with biotinylated anti–IFN-γ or anti–IL-4 antibody (clones 4S.B3 and MP4-25D2, respectively, from BD Pharmingen), and spots were counted under a Zeiss microscope using ELISPOT assay software. Negative controls were run in parallel using T cells without antigen, and the corresponding scores were subtracted from those of the unknowns.
Efficacy assessments.
As illustrated in Supplemental Figure 1, efficacy assessments included analysis of C7 expression and AFs in the injected skin compared with noninjected skin ~2 cm away, at 2 weeks, 3 months, and 12 months after treatment.
C7 IF.
Skin sections measuring 5 μm were air dried, fixed, and initially blocked with normal goat serum (Sigma-Aldrich) and then incubated with either mouse monoclonal anti-NC1 C7 antibody (primary antibody; clone LH 7.2; Sigma-Aldrich) diluted 1:1,000 in PBS or rabbit polyclonal anti-NC2 C7 antibody (primary antibody; ab198899; Abcam) diluted 1:500 in PBS, followed by detection with FITC-labeled goat anti-mouse secondary antibody (Invitrogen). All immunostaining experiments were repeated twice. Negative controls omitting the primary antibody were performed for each set of labeling experiments. All sections were photographed using the same camera and identical exposure times (1.5 seconds).
Quantification of MFI.
We measured MFI for 4 subjects (i.e., subject 1 excluded), in an assessor-blinded manner. The assessor quantifying the MFI received C7 IF images in 2 batches from the laboratory technician who processed the images, with each image coded by a random combination of letters and numbers. The assessor, blinded to the nature of the skin sections, then measured MFI using ImageJ, as previously described (30). Briefly, 10 measurements were taken at regular intervals using 8 × 8 pixels every 100 pixels along the DEJ. The statisticians were always unblinded. Mean values of the 10 measurements of C7 fluorescence intensity were calculated (giving C7 MFI), along with related SEM, for the injected and noninjected skin of the 4 subjects at 3 time points: week 2, month 3, and month 12, and the results from the injected skin were compared with respective results from noninjected skin and presented as a fold change and as absolute difference (12 comparisons in total). C7 MFI from normal human skin was used as a positive control.
TEM.
Skin biopsy specimens from both injected and noninjected sites were cut into small pieces (~1 mm3) and fixed overnight with half-strength Karnovsky’s fixative (2% [v/v] paraformaldehyde, 2.5% [v/v] glutaraldehyde in 0.1 M phosphate buffer [pH 7.4]) for 4 hours at room temperature and kept at 4°C until further processing. Skin samples were then rinsed several times in 0.1 M phosphate buffer and post-fixed with 1.3% osmium tetroxide (TAAB Laboratories) in double-distilled water for 2 hours at room temperature, followed by incubation in 2% uranyl acetate (Bio-Rad) in 50% ethanol and dehydrated in a graded series of ethanols. Tissue samples were further equilibrated with propylene oxide before infiltration with TAAB epoxy resin, embedded, and polymerized at 70°C for 24 hours. Ultrathin sections (70–90 nm) were prepared using a Reichert-Jung Ultracut E ultramicrotome mounted on 150 mesh copper grids (Gilder), contrasted using uranyl acetate and lead citrate, and examined in a JOEL JEM-1400+ transmission electron microscope. Images were acquired with an AMT 16000M camera (Advanced Microscopy Techniques Corp.).
Semiquantitative PCR of endogenous COL7A1 expression.
RNA was isolated from the injected and noninjected skin biopsies (week 2, month 3, and month 12), each measuring ~0.3 mm3, from subject 5 using the RNeasy Fibrous Tissue Mini Kit (QIAGEN) based on the manufacturer’s handbook, then treated with DNase (QIAGEN). First-strand cDNA synthesis was performed with 1 μg total RNA using Superscript II Reverse Transcriptase (Invitrogen). After cDNA was generated, semiquantitative PCRs were carried out using multiplex PCRs with GAPDH as internal control for each set of fragments of WT COL7A1 cDNA as follows: exon 54–56 forward 5′-CCTGGAAAAGCAGGCGAG-3′, reverse 5′-GGAGCCAGAGAGAAGGGAGA-3′; exon 86 forward 5′-GTGCCAGTGGAAAAGATGGA-3′; exon 87 forward 5′-CGGACCTAAAGGAGAACCTG-3′; and exon 90 reverse 5′-GTCCTCGGTCACCTTTGG-3′. First, the quantifiable PCR cycle was identified for each multiplex PCR from 24 identical PCR mixes with 1 μl cDNA together with patient-specific primers and GAPDH primer; 3 PCR tubes were put on ice at the end of alternate cycles from cycle 18 to 32 of the PCR program. The reactions were conducted in a Bio-Rad thermal cycler with initial denaturation at 95°C for 2 minutes, followed by 32 cycles of 95°C for 30 seconds, 58°C for 30 seconds, 72°C for 30 seconds, and a final extension at 72°C for 7 minutes. 1 μl of the PCR product from each tube was then used to run on 1.5% agarose gel by electrophoresis. Each band on the PCR gel image was used for quantification with GelQuant.NET (omicX).
PCR amplification of coCOL7A1 transgene.
Taking advantage of the difference between the cDNA sequence of codon-optimized COL7A1 transgene (coCOL7A1) and endogenous WT COL7A1, we designed primers specific for coCOL7A1 cDNA and performed PCR amplification to detect the presence of transgene in the injected skin samples of subject 5 at 2 weeks, 3 months, and 12 months after gene therapy using baseline sample as negative and transduced fibroblast cells as positive controls. Primers for coCOL7A1 cDNA were as follows: exon 12–13 forward 5′-AAAGAGAGACCTGCACCAG–3′, reverse 5′-TCTCTGATGGCTCTGATCAC-3′; exon 51–52 forward 5′-TGATAGCAGCGGCAGCTTTC-3′, reverse 5′-ATTCCAGGTTTTCCAGGCTCCC-3′; exon 112–113 forward 5′-CCGAGGAAGAGGAAAGAGTC-3′, reverse 5′-CCGAATCTATTGGCGTTGC-3′.
Sample size calculation.
RDEB is a rare disease, with an incidence of 3.05 per million live births (2). The study aimed to recruit 5–10 subjects as per the protocol. Even if no subjects experienced ARs (i.e., 0% AR), a sample size of 10 would give a 95% confidence interval of 0%–31% (exact CI was calculated using Stata software). A sample of 10 could still potentially provide reassurance that no more than 3 in 10 subjects should experience a given side effect. The trial steering committee approved the final study sample size of 4 patients enrolled as 5 subjects.
Statistics.
The mean C7 MFI and corresponding SEM were calculated from 10 measurements taken at regular intervals along the DEJ for the injected and noninjected sites, for each of subjects 2–5 at each time point (2 weeks, 3 months, and 12 months). Mean C7 MFI was compared between injected skin and noninjected skin separately for each of subjects 2–5, at each time point using a Mann-Whitney U test to allow for skewed data (12 comparisons in total). Each subject was analyzed separately due to large differences in baseline values of MFI, reflecting the different underlying subtypes of RDEB in these patients. The resulting significance test P values indicate whether the difference in C7 MFI between injected and noninjected sites (for a single subject at a single time point) is greater than would be expected by chance, given the variation in IF across the 10 sampled measurements. Statistically significant change was defined as P < 0.05. However, these 12 significance tests are post hoc and exploratory, and the P values should be interpreted with caution due to multiple testing. Full-length C7 MFI was similarly compared between injected and noninjected sites at each of the 3 time points, for subject 5 only, using the Mann-Whitney U test; again, this comparison is exploratory. Semiquantitative PCR of endogenous WT and mutant COL7A1 expression in subject 5 was performed in triplicate at 2 weeks, 3 months, and 12 months, and mean and SEM were calculated for each time point. Since these triplicate measurements essentially only provide a single “independent value” per time point (i.e., any variation between triplicates is due to measurement error only), statistical tests have not been applied to compare the COL7A1 expression values between the stated time points for subject 5.
Study approval.
This phase I clinical trial (EudraCT 2014-004884-19) was approved by the Medicines and Healthcare products Regulatory Agency (reference 14523/0258/001-0002) and Gene Therapy Advisory Committee (reference 15/LO/0665) in the United Kingdom in 2015. The study was registered at ClinicalTrials.gov (NCT02493816). The study and all investigations were conducted according to Declaration of Helsinki principles. Written informed consent was obtained from all patients prior to enrolment to the study, including the use of clinical photographs for publication.
Author contributions
SML, FS, WLD, JEM, WQ, and JAM contributed to protocol design; undertook the clinical trial; and contributed to data acquisition, analysis, and interpretation. SML, WLD, JEM, AJT, and JAM contributed to securing the grant. SML and JAM directed the overall study, analyzed data outcomes, and wrote the manuscript. AAW and MMQ contributed to protocol design and regulatory approvals, while TK and SS contributed to study undertaking and management. LL, AG, SA, PAL, JRM, JM, SM, MT, and AH contributed to acquisition and analysis of IF, TEM, ELISA, IIF, and ELISPOT data. FS, WLD, and AP contributed to the manufacture of the IMP, while LC and FF contributed to manufacture of GMP-grade lentiviral vectors. MC was the source of Chen antibodies for the Western blot analysis. FR, RP, and ME were the clinical trial statisticians and contributed to data analysis and presentation. FR also contributed to the study design. MDR, JT, JWB, and FL are members of the trial steering committee. MNA contributed to trial data analysis and interpretation. CG, LO, RR, CM, EO, ER, CT, and AO contributed to data acquisition and running of the trial. All authors contributed to manuscript review and approval of the final version.
Supplementary Material
Acknowledgments
Funding was received from Cure EB (previously known as Sohana Research Fund) in association with the Dystrophic Epidermolysis Bullosa Research Association (DEBRA, UK). The study was supported by the UK NIHR Biomedical Research Centres at Great Ormond Street, Guy’s and St Thomas’ NHS Foundation Trust, and King’s College London. The NIHR also supported WQ (RP2014). SML received a short-exchange fellowship from Fondation René Touraine to undertake the C7 ELISPOT assay at the Imagine Institute as part of this study. We are thankful to the EB clinical team, including EB specialist nurses at Guy’s and St Thomas’ NHS Foundation Trust and staff at the Clinical Research Facility, Guy’s Hospital, London, United Kingdom. Our special thanks go to all the patients for their time, enthusiasm, and dedicated commitment to participating in the trial and granting permission to publish this information.
Version 1. 06/06/2019
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019 American Society for Clinical Investigation
Reference information: JCI Insight. 2019;4(11):e126243. https://doi.org/10.1172/jci.insight.126243.
Contributor Information
Su M. Lwin, Email: su.m.lwin@kcl.ac.uk.
Farhatullah Syed, Email: farhatullah.syed@ucl.ac.uk.
Wei-Li Di, Email: w.di@ucl.ac.uk.
Tendai Kadiyirire, Email: Tendai.Kadiyirire@gstt.nhs.uk.
Lu Liu, Email: lu.liu@viapath.co.uk.
Alyson Guy, Email: alyson.guy@gstt.nhs.uk.
Anastasia Petrova, Email: a.petrova@ucl.ac.uk.
Alya Abdul-Wahab, Email: alyawahab@googlemail.com.
Fiona Reid, Email: fiona.reid@kcl.ac.uk.
Rachel Phillips, Email: r.phillips@imperial.ac.uk.
Maria Elstad, Email: maria.elstad@kcl.ac.uk.
Christos Georgiadis, Email: christos.georgiadis@ucl.ac.uk.
Sophia Aristodemou, Email: sophia.aristodemou@hotmail.com.
Patricia A. Lovell, Email: patricia.lovell@viapath.co.uk.
James R. McMillan, Email: jamesmcmillan57@hotmail.com.
John Mee, Email: john.mee@viapath.co.uk.
Snaigune Miskinyte, Email: snaigune.miskinyte@inserm.fr.
Matthias Titeux, Email: matthias.titeux@inserm.fr.
Linda Ozoemena, Email: linda.ozoemena@viapath.co.uk.
Rashida Pramanik, Email: rashida.pramanik@kcl.ac.uk.
Sonia Serrano, Email: sonia.serrano@gstt.nhs.uk.
Racheal Rowles, Email: rachealrowles@yahoo.co.uk.
Clarisse Maurin, Email: clarisse.maurin@free.fr.
Elizabeth Orrin, Email: elizabeth.orrin@nhs.net.
Magdalena Martinez-Queipo, Email: magdalena.martinez@gstt.nhs.uk.
Ellie Rashidghamat, Email: ellie.rashid@kcl.ac.uk.
Christos Tziotzios, Email: christos.tziotzios@kcl.ac.uk.
Alexandros Onoufriadis, Email: alexandros.onoufriadis@kcl.ac.uk.
Mei Chen, Email: chenm@usc.edu.
Lucas Chan, Email: lucas@cellvec.com.
Farzin Farzaneh, Email: farzin.farzaneh@kcl.ac.uk.
Marcela Del Rio, Email: mrnechae@ing.uc3m.es.
Jakub Tolar, Email: tolar003@umn.edu.
Johann W. Bauer, Email: jo.bauer@live.at.
Fernando Larcher, Email: fernando.larcher@ciemat.es.
Michael N. Antoniou, Email: michael.antoniou@kcl.ac.uk.
Alain Hovnanian, Email: alain.hovnanian@inserm.fr.
Adrian J. Thrasher, Email: a.thrasher@ucl.ac.uk.
Jemima E. Mellerio, Email: jemima.mellerio@kcl.ac.uk.
Waseem Qasim, Email: w.qasim@ucl.ac.uk.
John A. McGrath, Email: john.mcgrath@kcl.ac.uk.
References
- 1.Fine JD, Johnson LB, Weiner M, Li KP, Suchindran C. Epidermolysis bullosa and the risk of life-threatening cancers: the National EB Registry experience, 1986-2006. J Am Acad Dermatol. 2009;60(2):203–211. doi: 10.1016/j.jaad.2008.09.035. [DOI] [PubMed] [Google Scholar]
- 2.Fine JD. Epidemiology of inherited epidermolysis bullosa based on incidence and prevalence estimates from the National Epidermolysis Bullosa Registry. JAMA Dermatol. 2016;152(11):1231–1238. doi: 10.1001/jamadermatol.2016.2473. [DOI] [PubMed] [Google Scholar]
- 3.Fine JD, et al. Inherited epidermolysis bullosa: updated recommendations on diagnosis and classification. J Am Acad Dermatol. 2014;70(6):1103–1126. doi: 10.1016/j.jaad.2014.01.903. [DOI] [PubMed] [Google Scholar]
- 4.Christiano AM, et al. The large non-collagenous domain (NC-1) of type VII collagen is amino-terminal and chimeric. Homology to cartilage matrix protein, the type III domains of fibronectin and the A domains of von Willebrand factor. Hum Mol Genet. 1992;1(7):475–481. doi: 10.1093/hmg/1.7.475. [DOI] [PubMed] [Google Scholar]
- 5.Kirkorian AY, Weitz NA, Tlougan B, Morel KD. Evaluation of wound care options in patients with recessive dystrophic epidermolysis bullosa: a costly necessity. Pediatr Dermatol. 2014;31(1):33–37. doi: 10.1111/pde.12243. [DOI] [PubMed] [Google Scholar]
- 6.Woodley DT, et al. Injection of recombinant human type VII collagen restores collagen function in dystrophic epidermolysis bullosa. Nat Med. 2004;10(7):693–695. doi: 10.1038/nm1063. [DOI] [PubMed] [Google Scholar]
- 7.Woodley DT, et al. Intravenously injected recombinant human type VII collagen homes to skin wounds and restores skin integrity of dystrophic epidermolysis bullosa. J Invest Dermatol. 2013;133(7):1910–1913. doi: 10.1038/jid.2013.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Remington J, et al. Injection of recombinant human type VII collagen corrects the disease phenotype in a murine model of dystrophic epidermolysis bullosa. Mol Ther. 2009;17(1):26–33. doi: 10.1038/mt.2008.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wagner JE, et al. Bone marrow transplantation for recessive dystrophic epidermolysis bullosa. N Engl J Med. 2010;363(7):629–639. doi: 10.1056/NEJMoa0910501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tolar J, Wagner JE. Management of severe epidermolysis bullosa by haematopoietic transplant: principles, perspectives and pitfalls. Exp Dermatol. 2012;21(12):896–900. doi: 10.1111/exd.12014. [DOI] [PubMed] [Google Scholar]
- 11.Conget P, et al. Replenishment of type VII collagen and re-epithelialization of chronically ulcerated skin after intradermal administration of allogeneic mesenchymal stromal cells in two patients with recessive dystrophic epidermolysis bullosa. Cytotherapy. 2010;12(3):429–431. doi: 10.3109/14653241003587637. [DOI] [PubMed] [Google Scholar]
- 12.Petrof G, et al. Potential of systemic allogeneic mesenchymal stromal cell therapy for children with recessive dystrophic epidermolysis bullosa. J Invest Dermatol. 2015;135(9):2319–2321. doi: 10.1038/jid.2015.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McBride JD, Rodriguez-Menocal L, Badiavas EV. Extracellular vesicles as biomarkers and therapeutics in dermatology: a focus on exosomes. J Invest Dermatol. 2017;137(8):1622–1629. doi: 10.1016/j.jid.2017.04.021. [DOI] [PubMed] [Google Scholar]
- 14.Woodley DT, et al. Gentamicin induces functional type VII collagen in recessive dystrophic epidermolysis bullosa patients. J Clin Invest. 2017;127(8):3028–3038. doi: 10.1172/JCI92707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Titeux M, et al. SIN retroviral vectors expressing COL7A1 under human promoters for ex vivo gene therapy of recessive dystrophic epidermolysis bullosa. Mol Ther. 2010;18(8):1509–1518. doi: 10.1038/mt.2010.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sebastiano V, et al. Human COL7A1-corrected induced pluripotent stem cells for the treatment of recessive dystrophic epidermolysis bullosa. Sci Transl Med. 2014;6(264):264ra163. doi: 10.1126/scitranslmed.3009540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Osborn MJ, et al. TALEN-based gene correction for epidermolysis bullosa. Mol Ther. 2013;21(6):1151–1159. doi: 10.1038/mt.2013.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Droz-Georget Lathion S, et al. A single epidermal stem cell strategy for safe ex vivo gene therapy. EMBO Mol Med. 2015;7(4):380–393. doi: 10.15252/emmm.201404353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lwin SM, McGrath JA. Gene therapy for inherited skin disorders. eLS. 2017:1–15. [Google Scholar]
- 20.Goto M, et al. Fibroblasts show more potential as target cells than keratinocytes in COL7A1 gene therapy of dystrophic epidermolysis bullosa. J Invest Dermatol. 2006;126(4):766–772. doi: 10.1038/sj.jid.5700117. [DOI] [PubMed] [Google Scholar]
- 21.Jacków J, et al. Gene-corrected fibroblast therapy for recessive dystrophic epidermolysis bullosa using a self-inactivating COL7A1 retroviral vector. J Invest Dermatol. 2016;136(7):1346–1354. doi: 10.1016/j.jid.2016.02.811. [DOI] [PubMed] [Google Scholar]
- 22.Georgiadis C, et al. Lentiviral engineered fibroblasts expressing codon-optimized COL7A1 restore anchoring fibrils in RDEB. J Invest Dermatol. 2016;136(1):284–292. doi: 10.1038/JID.2015.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Di WL, et al. Phase I study protocol for ex vivo lentiviral gene therapy for the inherited skin disease, Netherton syndrome. Hum Gene Ther Clin Dev. 2013;24(4):182–190. doi: 10.1089/humc.2013.195. [DOI] [PubMed] [Google Scholar]
- 24.Wong T, et al. Potential of fibroblast cell therapy for recessive dystrophic epidermolysis bullosa. J Invest Dermatol. 2008;128(9):2179–2189. doi: 10.1038/jid.2008.78. [DOI] [PubMed] [Google Scholar]
- 25.Nagy N, et al. HB-EGF induces COL7A1 expression in keratinocytes and fibroblasts: possible mechanism underlying allogeneic fibroblast therapy in recessive dystrophic epidermolysis Bullosa. J Invest Dermatol. 2011;131(8):1771–1774. doi: 10.1038/jid.2011.85. [DOI] [PubMed] [Google Scholar]
- 26.Prost-Squarcioni C, et al. International Bullous Diseases Group: consensus on diagnostic criteria for epidermolysis bullosa acquisita. Br J Dermatol. 2018;179(1):30–41. doi: 10.1111/bjd.16138. [DOI] [PubMed] [Google Scholar]
- 27.Mavilio F, et al. Correction of junctional epidermolysis bullosa by transplantation of genetically modified epidermal stem cells. Nat Med. 2006;12(12):1397–1402. doi: 10.1038/nm1504. [DOI] [PubMed] [Google Scholar]
- 28.Hirsch T, et al. Regeneration of the entire human epidermis using transgenic stem cells. Nature. 2017;551(7680):327–332. doi: 10.1038/nature24487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Siprashvili Z, et al. Safety and wound outcomes following genetically corrected autologous epidermal grafts in patients with recessive dystrophic epidermolysis bullosa. JAMA. 2016;316(17):1808–1817. doi: 10.1001/jama.2016.15588. [DOI] [PubMed] [Google Scholar]
- 30.Pendaries V, et al. Immune reactivity to type VII collagen: implications for gene therapy of recessive dystrophic epidermolysis bullosa. Gene Ther. 2010;17(7):930–937. doi: 10.1038/gt.2010.36. [DOI] [PubMed] [Google Scholar]
- 31.Woodley DT, Briggaman RA, O’Keefe EJ, Inman AO, Queen LL, Gammon WR. Identification of the skin basement-membrane autoantigen in epidermolysis bullosa acquisita. N Engl J Med. 1984;310(16):1007–1013. doi: 10.1056/NEJM198404193101602. [DOI] [PubMed] [Google Scholar]
- 32.Myllylä R, et al. Expanding the lysyl hydroxylase toolbox: new insights into the localization and activities of lysyl hydroxylase 3 (LH3) J Cell Physiol. 2007;212(2):323–329. doi: 10.1002/jcp.21036. [DOI] [PubMed] [Google Scholar]
- 33.Sakai LY, Keene DR, Morris NP, Burgeson RE. Type VII collagen is a major structural component of anchoring fibrils. J Cell Biol. 1986;103(4):1577–1586. doi: 10.1083/jcb.103.4.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Compton CC, Gill JM, Bradford DA, Regauer S, Gallico GG, O’Connor NE. Skin regenerated from cultured epithelial autografts on full-thickness burn wounds from 6 days to 5 years after grafting. A light, electron microscopic and immunohistochemical study. Lab Invest. 1989;60(5):600–612. [PubMed] [Google Scholar]
- 35.da Costa Lima M, Jr, et al. Sensitivity of PCR and real-time PCR for the diagnosis of human visceral leishmaniasis using peripheral blood. Asian Pac J Trop Dis. 2013;3(1):10–15. doi: 10.1016/S2222-1808(13)60003-1. [DOI] [Google Scholar]
- 36.Bastien P, Procop GW, Reischl U. Quantitative real-time PCR is not more sensitive than “conventional” PCR. J Clin Microbiol. 2008;46(6):1897–1900. doi: 10.1128/JCM.02258-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krueger GG, et al. Genetically modified skin to treat disease: potential and limitations. J Invest Dermatol. 1994;103(5 Suppl):76S–84S. doi: 10.1111/1523-1747.ep12399100. [DOI] [PubMed] [Google Scholar]
- 38.Antoniou MN, Skipper KA, Anakok O. Optimizing retroviral gene expression for effective therapies. Hum Gene Ther. 2013;24(4):363–374. doi: 10.1089/hum.2013.062. [DOI] [PubMed] [Google Scholar]
- 39.Dighe N, et al. Long-term reproducible expression in human fetal liver hematopoietic stem cells with a UCOE-based lentiviral vector. PLoS One. 2014;9(8):e104805. doi: 10.1371/journal.pone.0104805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Woodley DT, et al. Normal and gene-corrected dystrophic epidermolysis bullosa fibroblasts alone can produce type VII collagen at the basement membrane zone. J Invest Dermatol. 2003;121(5):1021–1028. doi: 10.1046/j.1523-1747.2003.12571.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.