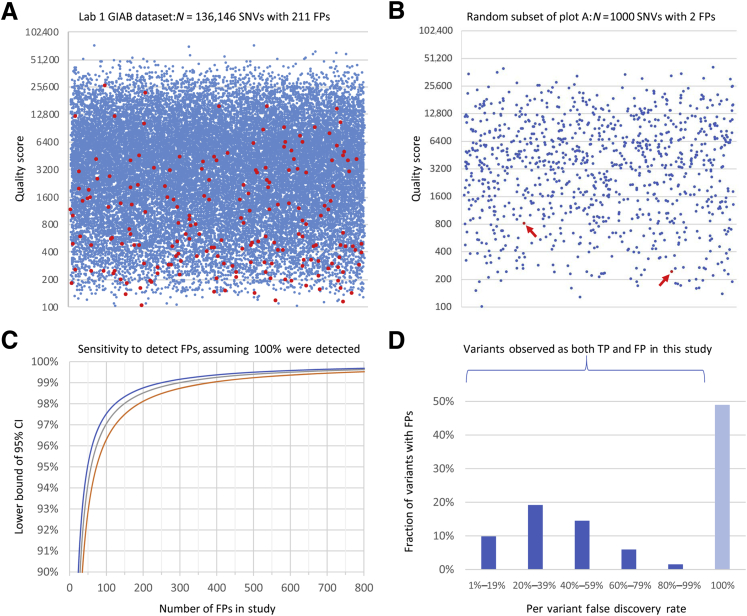
Figure 2.
Individual quality metric results and statistics. A: Variant call quality scores (QUAL, y axis) for true-positive (TP; blue) and false-positive (FP; red) single-nucleotide variant (SNV) calls in the Lab 1 Genome in a Bottle Consortium (GIAB) data set. The x axis position of each point is randomly assigned. To make density changes visible, a random selection of one-fifth of the TP calls was plotted along with all FPs. In the large data set, some FPs had quite high QUAL scores, demonstrating that this metric is inadequate alone. Corresponding histograms using the full data set without down-sampling are shown in Supplemental Figure S1. B: Random sample of 1000 data points from the same data set plotted in A. All points are displayed. Arrows indicate the two FPs present. One thousand such random samples were generated, and compared with the full data set in A, many would lead to quite different conclusions about the effectiveness of QUAL thresholds. C: Lower bound of the 95% CI on the fraction of FPs flagged (y axis) as a function of the number of FPs used to determine criteria (x axis) assuming that 100% success is observed. The y axis range is 90% to 100%. This calculation used the Jeffreys method (blue), the Wilson score method (red), and the tolerance interval method (gray). All methods produce generally similar results and indicate the validity of any study such as this. For example, using the Jeffreys method, flagging 49 of 49 FPs shows 100% effectiveness, with a CI of 95% to 100%. Many prior studies did not achieve this level of statistical significance (Table 1). Consistent with these CI calculations, small data sets indeed resulted in ineffective criteria (see Results). D: Histogram of per-variant false-discovery rates (FDRs; x axis) for all variants that were observed more than once in the Lab 1 data set, and for which one or more of those calls was an FP. SNVs and insertions and deletions (indels) are combined. An FDR of 100% indicates a fully systematic FP (insofar as we can measure); an FDR of 0% indicates a consistent TP (not shown in this graph). Each unique variant (ie, a genetic alteration that may be present in multiple individuals) is counted once. The y axis range is 0% to 50% of variants. Approximately half of all variants that were FPs were also correctly called as TPs in a different specimen(s) or run(s). Examples of this were observed in both the clinical and the GIAB specimens and included both SNVs and indels. Lab 2 results were similar. Many of these variants have low per-variant FDRs, which usually but not always are correctly called. Repeated TP observations of such a variant provide little information about the accuracy of any following observation of that same variant. This study was underpowered to measure FDRs near 0% or 100%, and many more of these variants may exist than are shown here.