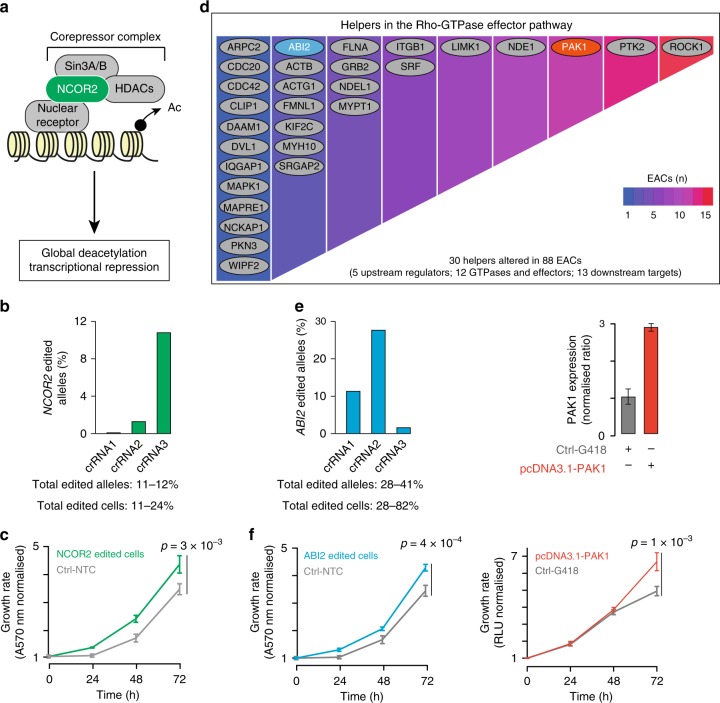
Fig. 5.
Cancer helper role of selected genes. a Function of NCOR2 as part of the nuclear receptor co-repressor complex, whose activity results in chromatin deacetylation and transcriptional repression. b Editing of the NCOR2 gene using three pooled crRNAs where cells are transiently co-transfected with Cas9 protein, crRNAs and tracrRNA32. The editing efficiency was measured using MiSeq. For NCOR2 two regions were sequenced, one containing the region targeted by crRNA1 and 2 and the other the region targeted bycrRNA3. For ABI2 only one amplicon was sequenced, containing the region targeted by all three crRNAs. The range of edited alleles and edited cells was derived considering the two opposite scenarios where all three crRNAs edit the same alleles/cells or different alleles/cells, respectively. c Proliferation curve of NCOR2 or non-targeting control (NTC) edited FLO-1 cells. d Manual curation of the helpers contributing to the Rho-GTPase effectors pathway. Heatmap indicates the number of samples with alterations in each gene. ABI2 (blue) and PAK1 (red) were selected for experimental validation. e Induced alterations in ABI2 and PAK1 genes. Editing of ABI2 was performed and assessed as described for NCOR2. PAK1 mRNA expression in FLO-1 cells was assessed by qRT-PCR, relativised to β-2-microglobulin and normalised to control cells. Experiments were done in triplicate in n = 2 biological replicates. f Proliferation curves of FLO-1 cells after ABI2 editing or PAK1 overexpression. For all proliferation assays, n = 3 biological replicates were performed, each with four technical replicates. Proliferation was assessed every 24 h and each time point was normalised to time zero. Mean values at 72 h were compared by two-tailed Student’s t-test. All error bars show standard deviation. Source data are available in the Source Data file