Figure 4.
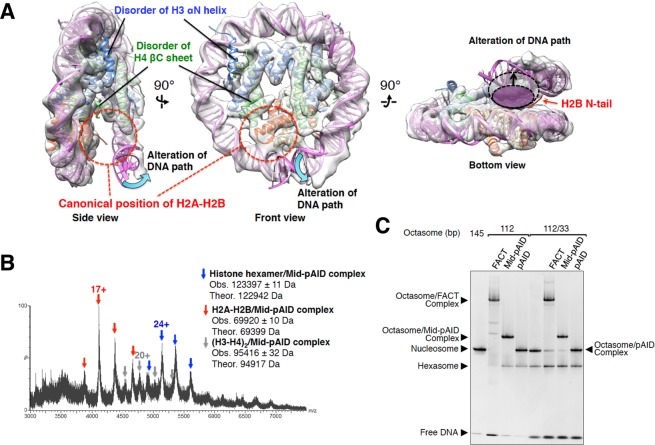
FACT causes partial conversion from octasome to hexasome. (A) Cryo-EM structure of the 112-bp hexasome. Three different views of the cryo-EM density map are superimposed onto the nucleosome structure (PDB ID: 2CV5), which lacks the 33-bp DNA and one H2A-H2B dimer. Red dotted circles show the canonical position of H2A-H2B in nucleosome (Side and Front views). Indicated structures of the H3 αN helix and the H4 βC sheet are disordered in the 112-bp hexasome map (Side and Front views). The cyan curved arrows show the significant alteration of the nucleosomal DNA path (Side and Front views). The dashed ellipsoids, shadowed in white and magenta, represent the space between two DNA gyres within the hexasome and octasome, respectively (Bottom view). The black arrows indicate the expanding distance between the hexasome and octasome (Bottom view). (B) Native ESI mass spectrum of the histone octamer mixed with Mid-pAID. Blue, red, and gray arrowheads show the corresponding peaks for the respective complexes, as indicated. The theoretical masses are calculated from those of the non-phosphorylated proteins. The charge states of the mainly observed peak are labeled above the peak. (C) EMSAs show hexasome formation upon binding of the FACT proteins to the 33/112-bp DSB nucleosome, detected by SYBR Gold nucleic acid gel stain. They are compared with the hexasome products in the 112-bp octasome reconstituted with the FACT proteins. Experiments were repeated at least three times.