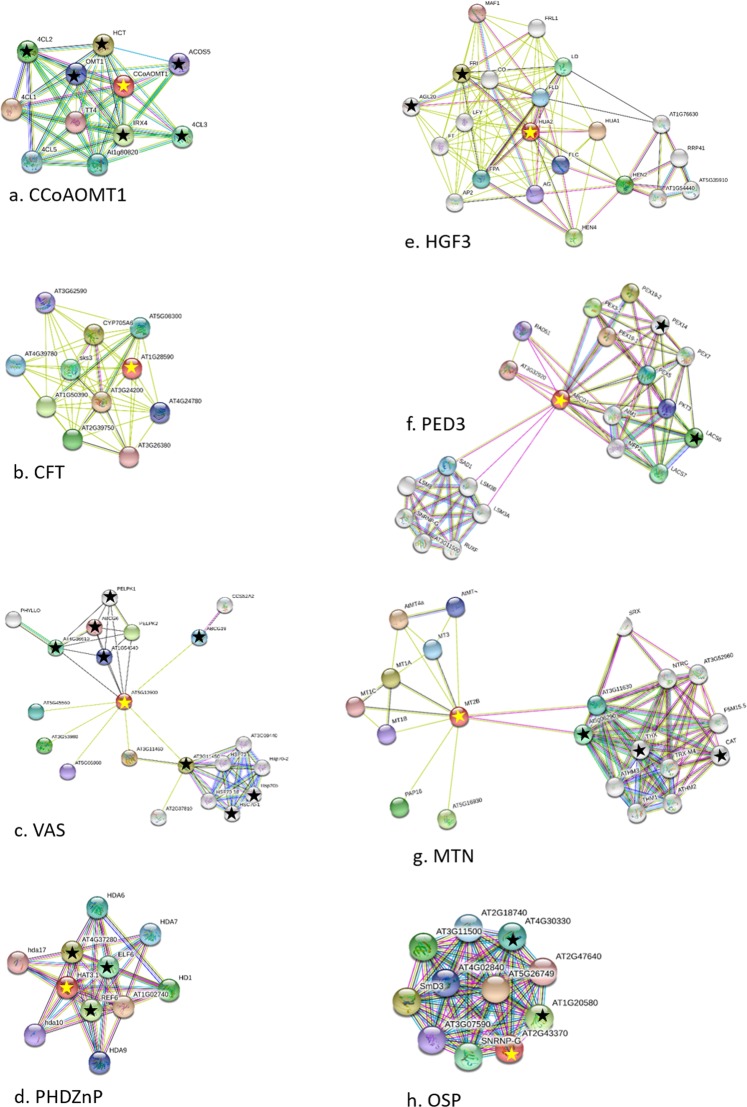
Figure 6.
STRING functional protein association networks (https://string-db.org/) of phellogen-related genes. Arabidopsis orthologs of the potato genes were identified and their TAIR IDs were used in the STRING search engine65. For each gene cluster, the query phellogen gene is marked with a yellow star and putative associated proteins that were also found in the potato phellogen transcriptome are marked with a black star. Data on the members of the interactomes is given in Table S1, Column Y, and in Table 1. In brief, (a) the interactome of CCoAOMT1 included phellogen-related genes, which are all involved in early stages of lignin biosynthesis. (b) The interactome of CFT included proteins of the GDSL esterase/lipase group that play a role in suberization, but none of those proteins were found in the phellogen transcriptome. (c) The interactome of VAS included phellogen proteins whose activities are related to heat stress-response and chromatin regulation, a cell-cycle regulator involved in the maintenance of shoot apical meristem, and ATP-binding cassettes which are involved in cell-wall synthesis. (d) The interactome of PHDZnP/HAT3 included the phellogen-related genes which are involved in the chromatin remodelling that controls meristem activity and organ polarity in an already established meristem. (e) The interactome of HGF3/HUA2 included the phellogen-related genes which are involved in specification of organ identity in meristem. (f) The interactome of PED3/CTS included the phellogen-related genes which are involved in transport and beta-oxidation of fatty acids and putatively with suberization. (g) The interactome of MTN included phellogen-related redox activities. (h) The interactome of OSP included phellogen-related small ribonuclear proteins that are involved in splicing of pre-mRNA and are required for plant development.