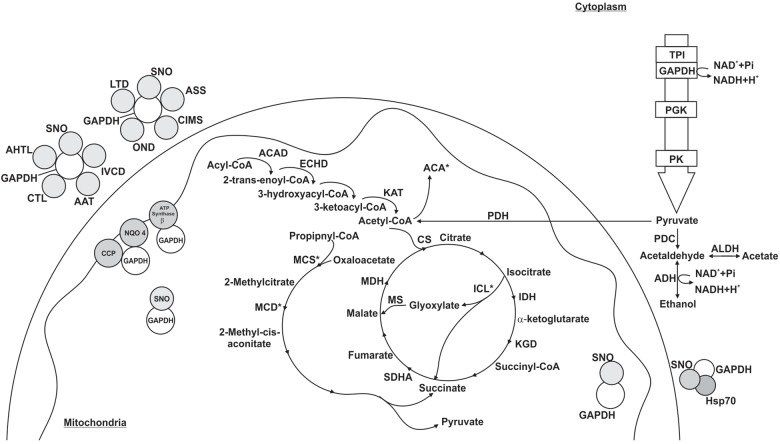
FIGURE 5.
Schematic model of the metabolic processes involving GAPDH through PPI in P. lutzii yeast phase. All proteins shown in the figures interacted with GAPDH. GAPDH (glyceraldehyde-3-phosphate dehydrogenase), SNO (nitrosylation), ACAD (acyl-CoA dehydrogenase), ECHD (enoyl-CoA hydratase), KAT (3-ketoacyl-CoA thiolase), CS (citrate synthase), IDH (isocitrate dehydrogenase), ICL (isocitrate lyase), KGD (dihydrolipoyl dehydrogenase), SDHA (succinate dehydrogenase flavoprotein subunit), MDH (malate dehydrogenase), MCS (2-methylcitrate synthase), MCD (2-methylcitrate dehydratase), TPI (triosephosphate isomerase), PGK (phosphoglycerate kinase), PK (pyruvate kinase), PDH (pyruvate dehydrogenase E1 component subunit β), PDC (pyruvate decarboxylase), ADH (alcohol dehydrogenase), ALDH (aldehyde dehydrogenase), ACA (acetyl-CoA acetyltransferase), AHTL (o-acetylhomoserine thiol-lyase), CTL (cystathionine γ-lyase), AAT (aspartate aminotransferase), IVCD (isovaleryl-CoA dehydrogenase), ASS (argininosuccinate synthase), CIMS (cobalamin-independent methionine synthase), OND (oxidoreductase 2-nitropropane dioxygenas), LTD (L-threonine 3-dehydrogenase), CCP (cytochrome c peroxidase) and NQO4 (NAD(P)H:quinone oxidoreductase type IV). The figure also shows the proteins HSP70 (heat shock protein 70) and ATP synthase β. Asterisks indicate proteins identified as GAPDH partners by both the chromatographic and BN-PAGE approaches.