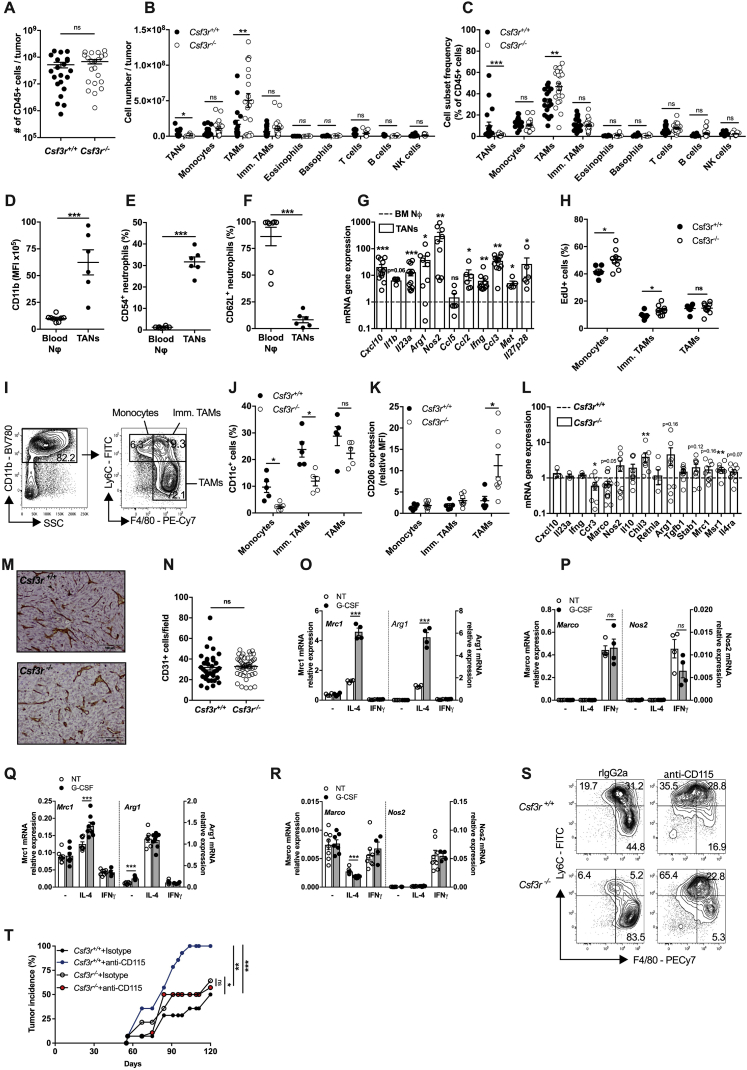
Figure S2.
TANs in Csf3r+/+ Mice Display an Activated Phenotype; Role of Macrophages in the Increased Susceptibility of Csf3r−/− to 3-MCA Sarcomagenesis, Related to Figures 1 and 2
(A-C) Number of sarcoma-infiltrating CD45+ cells (A), leukocyte cell subset frequencies (B) and absolute numbers (C) assessed by flow cytometry (tumor volume ≅ 2000 mm3).
(D-F) Quantification by flow cytometry of CD11b, CD54 and CD62L expression in TANs and peripheral-blood neutrophils from Csf3r+/+ sarcoma-bearing mice.
(G) mRNA gene expression in purified TANs. Gene expression was relative to Gapdh expression and normalized on the mean expression in naive bone marrow neutrophils (BM NΦ).
(H) Proliferative activity of tumor-infiltrating myeloid subsets, assessed by flow cytometry (intracellular EdU staining).
(I) Gating strategy for tumor-associated non-granulocytic myeloid populations. Cells represented in left dot plot are pregated on Aqua-/CD45+/Ly6G- cells.
(J-K) Flow cytometry analysis of CD206 and CD11c expression on tumor-associated non-granulocytic myeloid cells
(L) mRNA expression of M1- and M2-selected genes in tumor-infiltrating TAMs. Gene expression was relative to Gapdh expression and normalized on the mean of expression found in Csf3r+/+ TAMs.
(M-N) Immunohistochemical analysis and relative quantification of CD31+ cells in Csf3r+/+ and Csf3r−/− sarcomas. 5 random fields per sample were counted. Scale bar, 100 μm.
(O-R, mRNA expression of M1- and M2-related genes in BMDMs generated with M-CSF (O-P) or GM-CSF (Q-R) (see Methods). Gene expression was normalized on Gapdh expression.
(S) Representative dot plots showing depletion of TAMs after treatment with anti-CD115 antibody. (T) Incidence of 3-MCA-induced sarcomas in Csf3r+/+ and Csf3r−/− mice treated with anti-CD115 antibody or with isotype control.
(A-F), (H), (J-K), (N-R). Data are mean ± SEM. ∗p ≤ 0.05, ∗∗p ≤ 0.01 ∗∗∗p ≤ 0.001, ns, not statistically significant. (A), (D-F). Two-tailed Mann-Whitney U test. (B-C), (H), (J-K), (O-R) Two-tailed multiple Student’s t tests. (G), (L) Wilcoxon signed rank test. (T) Friedman test with Dunn’s multiple comparison test.
(A) n = 21 mice per group. (B) n = 6 Csf3r+/+ eosinophils, basophils, n = 8 Csf3r−/− eosinophils, basophils, n = 14 Csf3r+/+ T, NK cells, n = 9 Csf3r−/− T, NK cells, n = 9 Csf3r+/+ B cells, n = 6 Csf3r−/− B cells, n = 17 Csf3r+/+ TANs, monocytes, TAMs, immature TAMs, n = 20 Csf3r−/− TANs, monocytes, TAMs, immature TAMs. (C) n = 6 Csf3r+/+ eosinophils, basophils, n = 8 Csf3r−/− eosinophils, basophils, n = 18 Csf3r+/+ T, B, NK cells and TAMs, n = 19 TANs, monocytes, n = 20 Csf3r−/− B cells, n = 21 Csf3r−/− NK, T cells, n = 23 Csf3r−/− TANs, monocytes, immature TAMs, TAMs. (D-F) n = 6 TANs, n = 8 blood neutrophils. (G) n = 5 Il1b, Met, n = 6 Il27p28, Ccl5, Ccl2, n = 9 Nos2, Arg, Tnfa, n = 11 Ifng, Ccl3, n = 12 Cxcl10. (H) n = 6 (Csf3r+/+) or n = 9 (Csf3r−/−) mice. (J-K) n = 5 mice. (L) n = 3 (Cxcl10, Il23a, Ifng), n = 7 (Retnla), n = 8 (Chil3), n = 9 (Arg1, Stab1, Mrc1, Msr1, Il4ra), n = 10 (Ccr3, Nos2, Il10, Tgfb1). (M-N) n = 7 (Csf3r+/+) or n = 9 (Csf3r−/−) mice. (O-P) n = 4 mice per group. (Q-R), n = 8 (NT, IL-4, IL-4 + G-CSF, IFNγ), n = 7 (G-CSF), n = 4 (IFNγ+G-CSF). (H) n = 7 (Csf3r+/+ Isotype, Csf3r−/− Isotype, Csf3r−/− anti-CD115) or n = 8 (Csf3r+/+ anti-CD115) mice per group.
(A-C) Pooled data of four experiments are shown. (D-H), (M-N). (S-T) One experiment performed. (J-L) Pooled data from two (J-K) or three (L) experiments are shown. (O-R) Pooled data from two experiments.