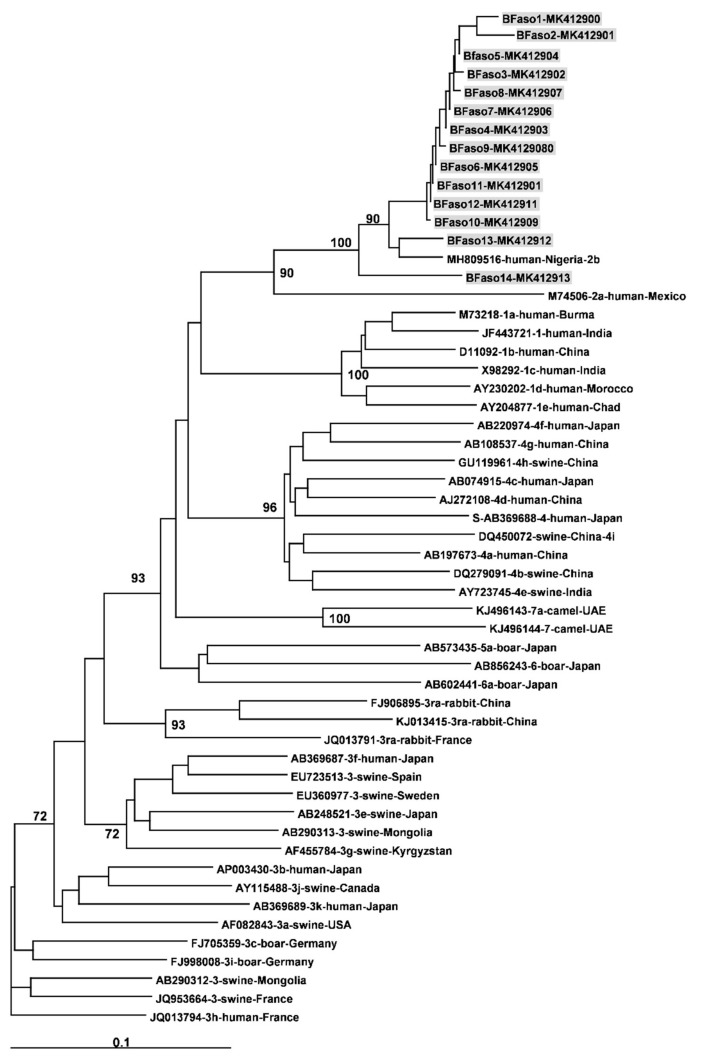
Figure 2.
Phylogenetic tree constructed using 347-nt-long partial sequences within ORF2 (black dots). Genetic distances were calculated using the Kimura two-parameter method, phylogenetic trees were plotted by the neighbour-joining method. Bootstrap values acquired after 100 replications are shown (branch lengths measured as the number of substitutions per site). Highlighted patient sequences (grey boxes) were compared to reference sequences (Smith et al., 2016) and to the new full length of subtype 2b strain (MH809516). Accession numbers, genotypes, and country of origin of collections are listed.