Abstract
Highly pathogenic avian influenza (HPAI) H5N1 and H5N8 have become endemic among domestic poultry in Egypt since 2006 and 2016, respectively. In parallel, the low pathogenic avian influenza H9N2 virus has been endemic since 2010. Despite the continuous circulation of these subtypes for several years, no natural reassortant has been detected so far among the domestic poultry population in Egypt. In this study, the HPAI (H5N2) virus was isolated from a commercial duck farm, giving evidence of the emergence of the first natural reassortment event in domestic poultry in Egypt. The virus was derived as a result of genetic reassortment between avian influenza viruses of H5N8 and H9N2 subtypes circulating in Egypt. The exchange of the neuraminidase segment and high number of acquired mutations might be associated with an alteration in the biological propensities of this virus.
Keywords: Avian influenza, H5N2, H5N8, H9N2, reassortment, poultry, Egypt
1. Introduction
Highly pathogenic avian influenza (HPAI) H5N1 and low pathogenic avian influenza (LPAI) H9N2 viruses have become endemic among domestic poultry in Egypt since 2006 and 2010, respectively [1]. In late 2016, HPAI H5N8 virus of clade 2.3.4.4 (group B) was first reported among wild birds in Egypt [2]. Since 2016, HPAI H5N8 viruses have been reported in different geographical regions across the country in both commercial and backyard bird sectors [3,4], where six genotypes have been detected in both wild and domestic birds [3,4,5]. Moreover, Egypt reported the highest number of human cases with HPAI H5N1 virus worldwide, and one of the only three countries (Egypt, China, and Bangladesh) that reported LPAI H9N2 in humans [1,6]. Recently, simultaneous detection of the three subtypes (H5N1, H5N8, and H9N2) has been described in a poultry farm in Egypt [7]. The co-circulation of those three subtypes increases risks for the generation of reassortants with unpredictable phenotypic properties, including an increased potential threat to human populations. Despite the co-circulation of HPAI H5N1 and LPAI H9N2 viruses for more than eight years, no natural reassortant has been detected so far among domestic poultry population in Egypt between those two subtypes. However, the risk of natural reassortant has been increased after the introduction of the HPAI H5N8 virus in 2016 [1]. This study reports and analyses the genetic and phylogenetic features of the first natural reassortant evidence in domestic poultry in Egypt.
2. Materials and Methods
2.1. Samples
On 31 December 2018, twenty oropharyngeal and cloacal swabs were collected from a commercial 90-days-old Muscovy duck farm (of 5000 birds) at Mansoura city, Dakahlia governorate, in Egypt. Samples were taken prior to slaughtering as a part of an active surveillance program conducted by the National Laboratory for veterinary quality control on poultry production (NLQP) and General Organization for Veterinary Services. Ducks were vaccinated via an H5N1 vaccine at 7 days old. Ducks were apparently healthy showing no signs of disease. Collected samples were submitted to NLQP for virus identification and isolation. All experiments in this study were conducted in accordance with the ethically approved protocol (AHRI-18032019) of the Animal Health Research Institute, Giza, Egypt.
2.2. RNA Extraction and Molecular Diagnosis
Viral RNA was extracted from the pooled samples using the QIAamp Viral RNA Mini Kit (Qiagen, Gmbh, Hilden, Germany) according to the manufacturer’s instructions. Pooled samples were tested using one step Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) (Qiagen, Gmbh, Hilden, Germany) for the M gene of influenza type A viruses [8] using the real-time PCR Mx3005P QPCR System (Agilent, Santa Clara, CA, USA). Positive avian influenza virus (AIV) RNA was subtyped for hemagglutinin (HA) and neuraminidase (NA) using specific subtyping RT-qPCR [9,10].
2.3. Virus Isolation
Virus isolation was performed via inoculation into the allantoic cavity of 10-day-old specific pathogen free (SPF) embryonated chicken eggs (ECE) according to the World Organization for Animal Health (OIE) diagnostic manual [11]. Collected allantoic fluid was tested using an hemaglutination assay and specific RT-qPCRs [9,10]. HA-positive allantoic fluids were stored at −80 °C.
2.4. Gene Sequencing
The complete genome sequences of the Egyptian virus A/duck/Egypt/VG1099/2018 (EG-VG1099) was amplified using RT-PCR with SuperScript-III One-Step RT-PCR System with Platinum® Taq DNA Polymerase (Invitrogen, Waltham, MA, USA) with primers described previously [12,13]. The gene-specific RT-PCR amplicons were size-separated using agarose gel electrophoresis, excised, and purified from gels using the QIAquick Gel Extraction Kit (Qiagen, Gmbh, Hilden, Germany). Further, purified PCR products were used directly for cycle sequencing reactions using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA). Reaction products were purified using Centrisep spin columns (ThermoFisher, Waltham, MA, USA) and sequenced on an ABI 3500XL Genetic Analyzer (Life Technologies, Carlsbad, CA, USA). Sequences generated in this study were submitted to the Global Initiative on Sharing All Influenza Data (GISAID) platform under accession number: EPI1387245-52.
2.5. Genetic and Phylogenetic Characterization
The obtained sequences were assembled and edited using the Geneious® software, version 11.0.5 [14]. A Basic Local Alignment Search Tool (BLAST) search was performed using Global Initiative on Sharing All Influenza Data (GISAID) platform, and sequences used in this study have been retrieved from the GISAID database for representative H5N8, H9N2, and other similar viruses. Alignment and identity matrix analyses were done using Multiple Alignment using Fast Fourier Transform (MAFFT) [15]. Phylogenetic analyses were based on a maximum likelihood methodology based on Bayesian (BIC) criterion after selection of the best fit models using IQ-tree software version 1.1.3 [16]. Trees were finally viewed and edited using FigTree v1.4.2 software (http://tree.bio.ed.ac.uk/software/figtree/).
3. Results
3.1. Virus Isolation
Pooled samples were found positive for the influenza A H5N2 subtype and negative for H7 and H9, as well as N1 and N8. Ducks were apparently healthy with neither disease symptoms nor abnormal mortalities. The virus was isolated, confirmed, and named A/duck/Egypt/VG1099/2018 (EG-VG1099).
3.2. Genetic and Phylogenetic Characterization
The complete genome sequences of the Egyptian virus EG-VG1099 were obtained and sequences generated in this study were submitted to the Global Initiative on Sharing All Influenza Data (GISAID) platform under the accession number: EPI1387245-52. Nucleotide identity analysis showed that the EG-VG1099 virus was closely related, according to its hemagglutinin (HA) gene segment, to HPAI H5N8 viruses isolated in Europe 2016–2017, with a nucleotide sequence identity of 98% to the A/Eur_Wig/NL-Greonterp/16015653-001/2016; the same identity of 98% with A/duck/Egypt/F446/2017(H5N8) virus was also recorded. The neuraminidase (NA) gene of the EG-VG1099 virus shared 97% nucleotide identity with the Egyptian LPAI H9N2 virus A/chicken/Egypt/D10700/2015(H9N2) (Table 1), and no identity with any virus outside Egypt in the first 200 hits was observed. Six segments—polymerase basic (PB2), PB1, polymerase acidic (PA), nucleoprotein (NP), matrix protein (M), nonstructural protein (NS)—revealed a nucleotide identity of 98%–99% with the Dutch (A/Eur_Wig/NL-Greonterp/16015653-001/2016) and Italian (A/swan/Italy/17VIR537-2/2017) HPAI H5N8 viruses (Table 1). Possible reassortment option of the novel Egyptian HPAI H5N2 virus is illustrated in Figure 1C.
Table 1.
Nucleotide sequence identities between the EG-VG1099 (H5N2) virus and nearest homologues in the GenBank and GISAID database.
| Gene | Virus | Subtype | Accession Number | Collection Date | Identity |
|---|---|---|---|---|---|
| PB2 | A/Ruddy Shelduck/AN/2-14-12/2016 | H5N8 | EPI1184014 | 2016-12-14 | 98% |
| A/swan/Italy/17VIR537-2/2017 | H5N8 | EPI954556 | 2017-01-19 | 98% | |
| A/chicken/Egypt/Gharbiya-15/2017 | H5N8 | EPI1104288 | 2017 | 98% | |
| A/Eur_Wig/NL-Greonterp/16015653-001/2016 | H5N8 | EPI1019555 | 2016-12-08 | 98% | |
| Pb1 | A/White-fronted Goose/AN/1-15-12/2016 | H5N8 | EPI1184005 | 2016-12-15 | 99% |
| A/Eur_Wig/NL-Greonterp/16015653-001/2016 | H5N8 | EPI1019556 | 2016-12-08 | 99% | |
| A/duck/Egypt/F446/2017 | H5N8 | EPI1018205 | 2017-04-06 | 99% | |
| A/swan/Italy/17VIR537-2/2017 | H5N8 | EPI954557 | 2017-01-19 | 99% | |
| PA | A/Eur_Wig/NL-Greonterp/16015653-001/2016 | H5N8 | EPI1019554 | 2016-12-08 | 98% |
| A/White-fronted Goose/AN/1-15-12/2016 | H5N8 | EPI1184004 | 2016-12-15 | 98% | |
| A/duck/Egypt/F446/2017 | H5N8 | EPI1018206 | 2017-04-06 | 98% | |
| A/swan/Italy/17VIR537-2/2017 | H5N8 | EPI954555 | 2017-01-19 | 98% | |
| HA | A/Eur_Wig/NL-Greonterp/16015653-001/2016 | H5N8 | EPI1019558 | 2016-12-08 | 98% |
| A/swan/Italy/17VIR537-2/2017 | H5N8 | EPI954559 | 2017-01-19 | 98% | |
| A/White-fronted Goose/AN/1-15-12/2016 | H5N8 | EPI1183999 | 2016-12-15 | 98% | |
| A/duck/Egypt/F446/2017 | H5N8 | EPI1018207 | 2017-04-06 | 98% | |
| NP | A/chicken/Egypt/Gharbiya-15/2017 | H5N8 | EPI1104284 | 2017 | 99% |
| A/duck/Egypt/F446/2017 | H5N8 | EPI1018208 | 2017-04-07 | 99% | |
| A/Eur_Wig/NL-Greonterp/16015653-001/2016 | H5N8 | EPI1019551 | 2016-12-08 | 99% | |
| A/swan/Italy/17VIR537-2/2017 | H5N8 | EPI954552 | 2017-01-19 | 99% | |
| NA | A/chicken/Egypt/D10700/2015 | H9N2 | KX000734* | 2015-02-26 | 97% |
| A/chicken/Egypt/S107569A/2015 | H9N2 | KX000727* | 2015-02-15 | 97% | |
| A/chicken/Egypt/A1093D/2015 | H9N2 | KX000717* | 2015-02-09 | 97% | |
| M | A/chicken/Egypt/Gharbiya-15/2017 | H5N8 | EPI1104286 | 2017 | 98% |
| A/duck/Egypt/F446/2017 | H5N8 | EPI1018210 | 2017-04-06 | 98% | |
| A/swan/Italy/17VIR537-2/2017 | H5N8 | EPI954554 | 2017-01-19 | 98% | |
| A/Eur_Wig/NL-Greonterp/16015653-001/2016 | H5N8 | EPI1019553 | 2016-12-08 | 98% | |
| NS | A/Eur_Wig/NL-Greonterp/16015653-001/2016 | H5N8 | EPI1019552 | 2016-12-08 | 98% |
| A/chicken/Egypt/Gharbiya-15/2017 | H5N8 | EPI1104285 | 2017 | 98% | |
| A/swan/Italy/17VIR537-2/2017 | H5N8 | EPI954553 | 2017-01-19 | 98% | |
| A/duck/Egypt/F446/2017 | H5N8 | EPI1018211 | 2017-04-06 | 98% |
* GenBank accession number.
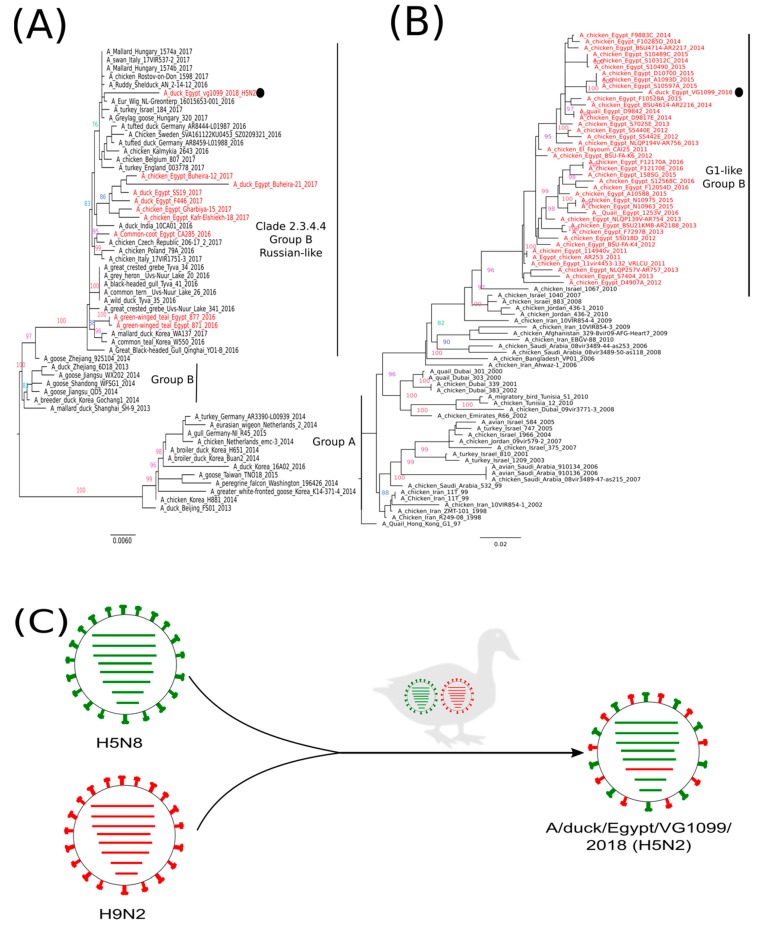
Figure 1.
Phylogenetic trees of the nucleotide sequences of the HA (A) and NA (B) gene segments. Maximum likelihood calculations were done using the IQTree software under the best fit model according to the Bayesian criterion. Egyptian H5N2 virus is indicated with a black dot. (C) Emergence and possible reassortment option of the novel HPAI H5N2 virus found in Egypt in 2018.
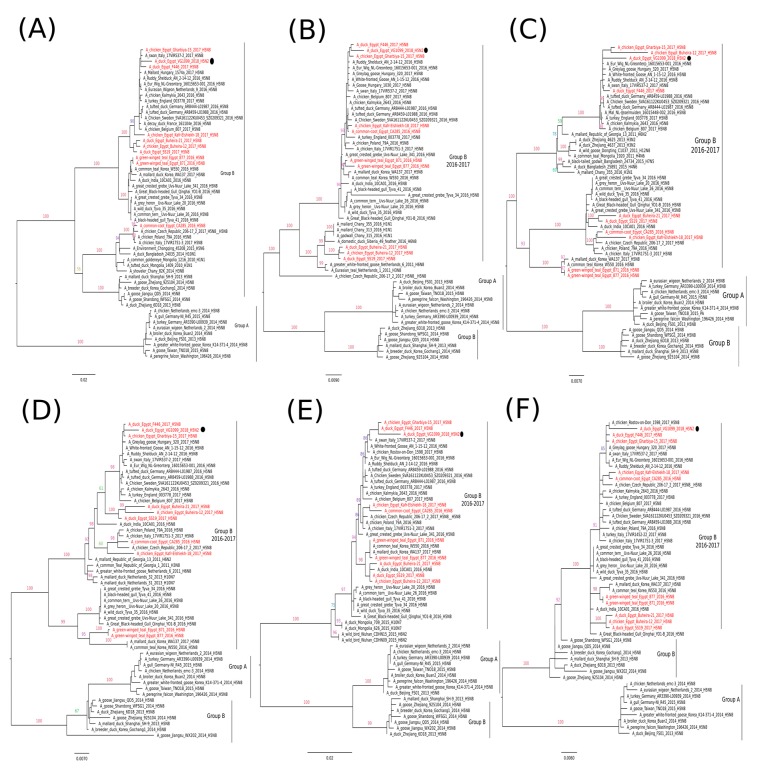
Phylogenetic analysis of the HA gene segment showed that the EG-VG1099 virus was closely related to HPAI H5N8 viruses reported in Europe during 2016–2017 (2.3.4.4 clade, group B), and phylogenetically distinguished from the previously reported HPAI H5N8 viruses in Egypt (Figure 1A). The NA gene segment of EG-VG1099 was phylogenetically grouped with the LPAI H9N2 viruses isolated from Egypt (Figure 1B). The remaining six genes (PB2, PB1, PA, NP, M, and NS) of the EG-VG1099 virus were clustered with Egyptian HPAI H5N8 viruses: A/chicken/Egypt/Gharbiya-15/2017 and A/duck/Egypt/F446/2017, and revealed a close phylogenetic relatedness with H5N8 viruses frequently found in Europe in 2016–2017 (Figure 2).
Figure 2.
Phylogenetic trees of the nucleotide sequences of the gene segments PB2, PB1, PA, NP, M, and NS (A–F). Maximum likelihood calculations were done with the IQTree software under the best fit model according to the Bayesian criterion. Egyptian HPAI H5N8 viruses are colored in red and the H5N2 virus is indicated with a black dot.
The HA gene of the EG-VG1099 virus possessed multiple basic amino acids, “PLREKRRKR/GLF”, in the cleavage site of the HA indicating high pathogenicity of this virus. The EG-VG1099 virus exhibited R76S, S98R, A138S, A160V, and R173Q amino acid substitutions (H3 numbering) at its HA protein, distinguishing it from previously reported HPAI H5N8 viruses of clade 2.3.4.4 (group B). Amino acid substitutions at positions S98R, A138S, and A160V have been reported previously to be related to virulence and host specificity [17,18]. The NA coded protein of the emerged HPAI H5N2 virus was distinguished by 43 nucleotide mutations from the Egyptian LPAI H9N2 viruses (during 2010–2016, no available published sequence hereafter). Twelve out of the 43 were non-synonymous and encoded amino acid substitutions: F37L, T43A, V50E, K143E, R199K, M210I, L211I, R283Q, R288I, R344V, N384K, and K415R. Positions 143 and 344 were located in the antibodies recognition sites of the NA coded protein [19,20]. However, none of the remaining amino acid substitutions have been described in association with resistance to neuraminidase inhibitors or located in the hemadsorption site. Further, the Egyptian EG-VG1099 possessed no mutations at known molecular features associated with virulence or host adaptation, like E627K and D701N in PB2, N375S and L598P in PB1, or V100A in PA [17]. However, N30D and T215A amino acid substitutions in the M1 protein and P42S in the NS1 protein (previously reported in the Egyptian H5N8 viruses) were observed, suggesting that the virus could exhibit increased virulence in mammals [17].
4. Discussion
Currently, Egypt faces endemic co-circulation of HPAI (H5N1, H5N8) and LPAI (H9N2) viruses, where a simultaneous detection of the three subtypes has been detected [7]. The co-circulation of those three subtypes raises fears for the generation of a new subtype/genotype with unpredictable properties, including an increased potential threat to human [1]. However, the nucleotide identity and phylogenetic distance of the Egyptian HPAI H5N2 virus (EG-VG1099) virus with other Egyptian HA genes seems low, and it is reasonable to suggest that the new reassortant EG-VG1099 resulted from a reassortant between HPAI H5N8 virus of clade 2.3.4.4 (group B) and the Egyptian LPAI (H9N2) virus of the G1-like lineage (Figure 1C). This was supported by the phylogenetic relatedness of the NA gene segment with only viruses isolated from Egypt; however, the circulation of an additional variant of H5N8 in Egypt cannot be excluded. So far, no natural reassortant has been detected between the Egyptian H5N1/H9N2 subtypes [21], and the emerged H5N2 virus in this study indicated a higher reassortment compatibility between the Egyptian H5N8 and H9N2 viruses compared to the Egyptian H5N1/H9N2. However, we cannot exclude the detection of new genotypes with different gene constellations in the upcoming period. This highlights the significance of obtaining the whole genome sequence of the circulating HPAI H5N1/H5N2/H5N8 viruses in Egypt.
Further, different amino acid substitutions associated with either virulence or host adaptation have been observed in the newly detected Egyptian HPAI H5N2 virus (EG-VG1099). Amino acid substitutions (S98R, A138S, and A160V) in the antibody recognition sites of the HA have been observed. These mutations were described to enhance the binding of AIV H5 with the α2.6 sialic acid receptor and increase virulence in the mammalian animal model [17,18]. Further, N30D and T215A amino acid substitutions in the M1 protein of the EG-VG1099 virus were reported to enhance the virulence of the HPAI H5N1 virus in mice [17]. In addition, P42S in the NS1 protein also has been found to increase the virulence of the HPAI H5N1 virus in mice [17]. Interestingly, a high number of non-synonymous nucleotide mutations (n = 12) in the NA have been noticed. Only positions 143 and 344 are located in the antibody recognition sites of the NA coded protein and have been reported in the literature to be related to the antigenic drift/escape mutant [19,20]. However, none of the rest have been described in association with resistance to neuraminidase inhibitors or located in the hemadsorption site. At this point, it cannot be excluded that the new HPAI H5N2 subtype, reported in this study, may be associated with an alternation in the biological propensities. Hence, future studies in birds (e.g., chickens, ducks) and mammalian models (e.g., ferrets, mice) is recommended to provide experimental data on the fitness and virulence of this virus subtype. In addition, in vivo and in vitro studies comparing the newly detected H5N2 virus to a closely related H5N8 virus are required to determine any alteration in the phenotypic properties between the two viruses.
Moreover, our study underlines the importance of active surveillance in the timely detection of new AIV subtypes. It can be expected that continuing surveillance activities might lead to the detection of additional new cases; hence, surveillance should be strengthened with special emphasis in the surrounding regions of the outbreak. Interventions, like improved biosecurity practices in poultry production enterprises, and live bird trading and marketing practices in Egypt, should be considered to reduce the dissemination of AIV and reduce the chance of reassortment raised through co-infection.
Acknowledgments
We also acknowledge the Food and Agriculture Organization of the United Nations (FAO), and GOVs Egypt for their continuous support in avian influenza surveillance programs. We thank Rachel Hickman for English language editing. We are grateful to the data submitters for their sequences in the GISAID database.
Author Contributions
Conceptualization, M.M.N., M.K.H., A.M.M.A., M.A., E.I., W.M.M.H and M.A.S. (Momtaz A. Shahein); methodology, N.M.H., A.M.E., A.G.S., M.E.-H., M.A.S. (Mohamed A. Saif), M.M.T., A.A.N., H.A.F. and A.A.S.; formal analysis, M.M.N.; investigation, M.E.-H., N.M.H. and M.M.N.; data curation, M.E.-H., M.M.N.; writing—original draft preparation, M.M.N.; writing—review and editing, A.M.E., N.M.H., A.-S.A. and M.M.N.
Funding
The authors acknowledge the Egyptian Ministry of Agriculture for funding this work. The work was funded in part by the FAO and USAID. This work was supported in part by the Swedish Research Council VR (grants number 2018-02569) to M.M.N.
Conflicts of Interest
Authors declare no conflict of interest.
References
- 1.Naguib M.M., Harder T. Endemic situation of multiple avian influenza strains in poultry in Egypt: A continuing nightmare. Zoonoses Public Health. 2018;65:908–910. doi: 10.1111/zph.12486. [DOI] [PubMed] [Google Scholar]
- 2.Selim A.A., Erfan A.M., Hagag N., Zanaty A., Samir A.H., Samy M., Abdelhalim A., Arafa A.A., Soliman M.A., Shaheen M., et al. Highly Pathogenic Avian Influenza Virus (H5N8) Clade 2.3.4.4 Infection in Migratory Birds, Egypt. Emerg. Infect. Dis. 2017;23:1048–1051. doi: 10.3201/eid2306.162056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salaheldin A.H., El-Hamid H.S., Elbestawy A.R., Veits J., Hafez H.M., Mettenleiter T.C., Abdelwhab E.M. Multiple Introductions of Influenza A(H5N8) Virus into Poultry, Egypt, 2017. Emerg. Infect. Dis. 2018;24:943–946. doi: 10.3201/eid2405.171935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yehia N., Naguib M.M., Li R., Hagag N., El-Husseiny M., Mosaad Z., Nour A., Rabea N., Hasan W.M., Hassan M.K., et al. Multiple introductions of reassorted highly pathogenic avian influenza viruses (H5N8) clade 2.3.4.4b causing outbreaks in wild birds and poultry in Egypt. Infect. Genet. Evol. 2018;58:56–65. doi: 10.1016/j.meegid.2017.12.011. [DOI] [PubMed] [Google Scholar]
- 5.Kandeil A., Kayed A., Moatasim Y., Webby R.J., McKenzie P.P., Kayali G., Ali M.A. Genetic characterization of highly pathogenic avian influenza A H5N8 viruses isolated from wild birds in Egypt. J. Gen. Virol. 2017;98:1573–1586. doi: 10.1099/jgv.0.000847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.WHO Cumulative Number of Confirmed Human Cases for Avian Influenza A(H5N1) Reported to WHO, 2003–2019. [(accessed on 24 March 2019)]; Available online: https://www.who.int/influenza/human_animal_interface/2019_02_12_tableH5N1.pdf?ua=1.
- 7.Shehata A.A., Sedeik M.E., Elbestawy A.R., Zain El-Abideen M.A., Ibrahim H.H., Kilany W.H., Ali A. Co-infections, genetic, and antigenic relatedness of avian influenza H5N8 and H5N1 viruses in domestic and wild birds in Egypt. Poult. Sci. 2019;98:2371–2379. doi: 10.3382/ps/pez011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spackman E., Senne D.A., Bulaga L.L., Myers T.J., Perdue M.L., Garber L.P., Lohman K., Daum L.T., Suarez D.L. Development of real-time RT-PCR for the detection of avian influenza virus. Avian Dis. 2003;47:1079–1082. doi: 10.1637/0005-2086-47.s3.1079. [DOI] [PubMed] [Google Scholar]
- 9.Monne I., Ormelli S., Salviato A., de Battisti C., Bettini F., Salomoni A., Drago A., Zecchin B., Capua I., Cattoli G. Development and validation of a one-step real-time PCR assay for simultaneous detection of subtype H5, H7, and H9 avian influenza viruses. J. Clin. Microbiol. 2008;46:1769–1773. doi: 10.1128/JCM.02204-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoffmann B., Hoffmann D., Henritzi D., Beer M., Harder T.C. Riems influenza a typing array (RITA): An RT-qPCR-based low density array for subtyping avian and mammalian influenza a viruses. Sci. Rep. 2016;6:27211. doi: 10.1038/srep27211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.OIE Chapter 3.3.4. Avian Influenza. [(accessed on 24 March 2019)]; Available online: http://www.oie.int/fileadmin/Home/eng/Health_standards/tahm/3.03.04_AI.pdf.
- 12.Hoper D., Hoffmann B., Beer M. Simple, sensitive, and swift sequencing of complete H5N1 avian influenza virus genomes. J. Clin. Microbiol. 2009;47:674–679. doi: 10.1128/JCM.01028-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Naguib M.M., Arafa A.S., El-Kady M.F., Selim A.A., Gunalan V., Maurer-Stroh S., Goller K.V., Hassan M.K., Beer M., Abdelwhab E.M., et al. Evolutionary trajectories and diagnostic challenges of potentially zoonotic avian influenza viruses H5N1 and H9N2 co-circulating in Egypt. Infect. Genet. Evol. 2015;34:278–291. doi: 10.1016/j.meegid.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 14.Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C., et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nguyen L.T., Schmidt H.A., von Haeseler A., Minh B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol. Biol. Evol. 2014;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.CDC, H5N1 Genetic Changes Inventory: A Tool for Influenza Surveillance and Preparedness. [(accessed on 24 March 2019)]; Available online: https://www.cdc.gov/flu/pdf/avianflu/h5n1-inventory.pdf.
- 18.De Graaf M., Fouchier R.A. Role of receptor binding specificity in influenza A virus transmission and pathogenesis. EMBO J. 2014;33:823–841. doi: 10.1002/embj.201387442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gulati U., Hwang C.C., Venkatramani L., Gulati S., Stray S.J., Lee J.T., Laver W.G., Bochkarev A., Zlotnick A., Air G.M. Antibody epitopes on the neuraminidase of a recent H3N2 influenza virus (A/Memphis/31/98) J. Virol. 2002;76:12274–12280. doi: 10.1128/JVI.76.23.12274-12280.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Air G.M., Els M.C., Brown L.E., Laver W.G., Webster R.G. Location of antigenic sites on the three-dimensional structure of the influenza N2 virus neuraminidase. Virology. 1985;145:237–248. doi: 10.1016/0042-6822(85)90157-6. [DOI] [PubMed] [Google Scholar]
- 21.Kim S.-H. Challenge for One Health: Co-Circulation of Zoonotic H5N1 and H9N2 Avian Influenza Viruses in Egypt. Viruses. 2018;10:121. doi: 10.3390/v10030121. [DOI] [PMC free article] [PubMed] [Google Scholar]