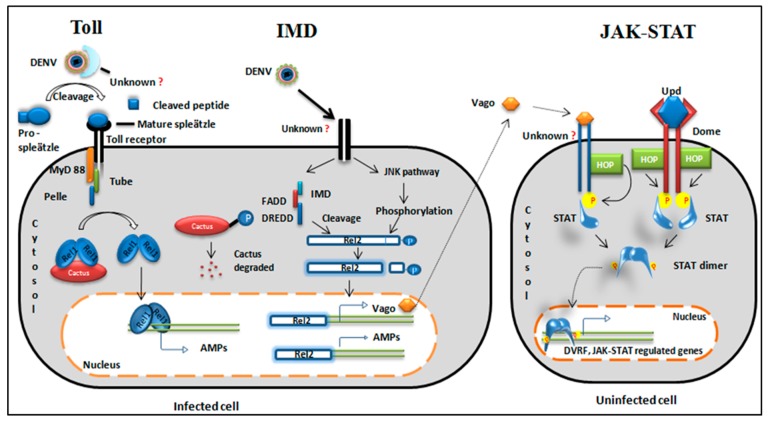
Figure 2.
Evolutionarily conserved signaling pathways. Toll pathway: The virus is recognized by an unknown cytoplasmic receptor. Cytokine pro-spleatzle is cleaved to active cytokine spleatzle and binds to the Toll receptor. Adaptor proteins are recruited to the Toll receptor. A negative regulator of cactus degrades and a free Rel1 dimer translocates to the nucleus. The Rel1 dimer acts as a transcription factor for Toll regulated genes and produces antimicrobial peptides (AMPs) (cecropin and defensin). IMD pathway: The virus binds to a transmembrane receptor of the cell and splits the pathway into two segments. One segment activates the JNK pathway and phosphorylates Rel2. Another segment recruits IMD and other adaptor proteins {Immune deficiency (IMD), fas-associated death domain (FADD) and death related ced-3/Nedd2-like protein (DREDD)} to cleave the C-terminal phosphorylated domain of Rel2.11. Cleaved Rel2 translocates to the nucleus and transcribes AMPs and a secretory protein, Vago. JAK/STAT pathway: This pathway is activated either by cytokine-like secretory protein from an infected cell or by the conserved JAK-STAT ligand Upd. Vago is secreted from nearby infected cells, binds to an unknown receptor, and recruits hopscotch (HOP) kinase. Similarly, Dome receptors bind Upd and receptor phosphorylation occurs through HOP kinase. The phosphorylated receptor is a docking site for STAT. STAT phosphorylation leads to dimerization. STAT dimer translocates to the nucleus and transcribes JAK/STAT-regulated genes and dengue virus restriction factor (DVRF).