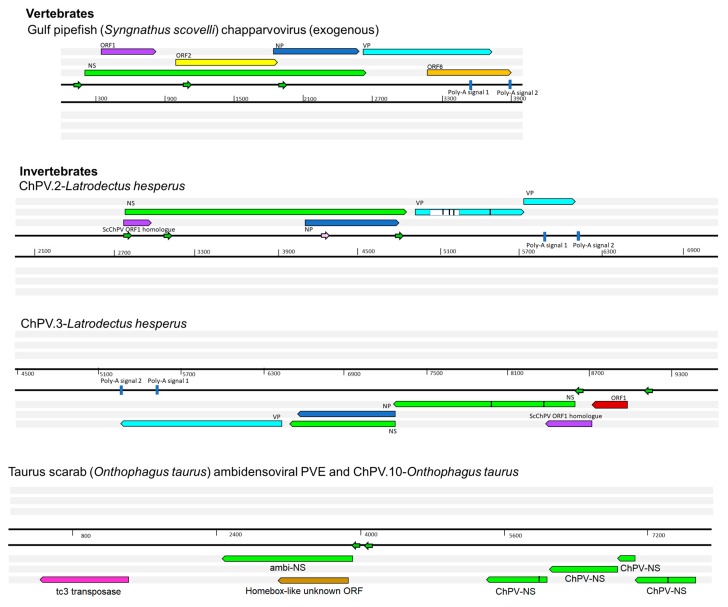
Figure 5.
Genomic structures of newly identified chapparvoviruses (ChPVs) and ChPV-derived endogenous parvoviral element (EPV) sequences. The positions of putative open reading frames (ORFs) and predicted cis transcription elements of ChPVs are shown. ChPV.2-LatHes contains a previously unidentified repetitive element, present as multiple copies scattered in the Latrodectus genome, marked by the white box within the VP gene. The element ChPV.10-OntTau shares its integration site with another endogenous element, disclosing similarity to ambidensoviruses. ORFs are represented by rectangular arrows, colored according to homology. In-frame stop codons are shown as vertical lines. Splice donor sites are marked by white-colored bars, acceptor sites by orange-colored bars. Blue-colored bars show predicted polyadenylation signals. Small arrows show predicted promoters and are colored according to prediction score (>0.95 = green; 0.9–0.95 = pink: <0.9 = yellow). Grey boxes indicate regions inferred to be transcribed but not translated.