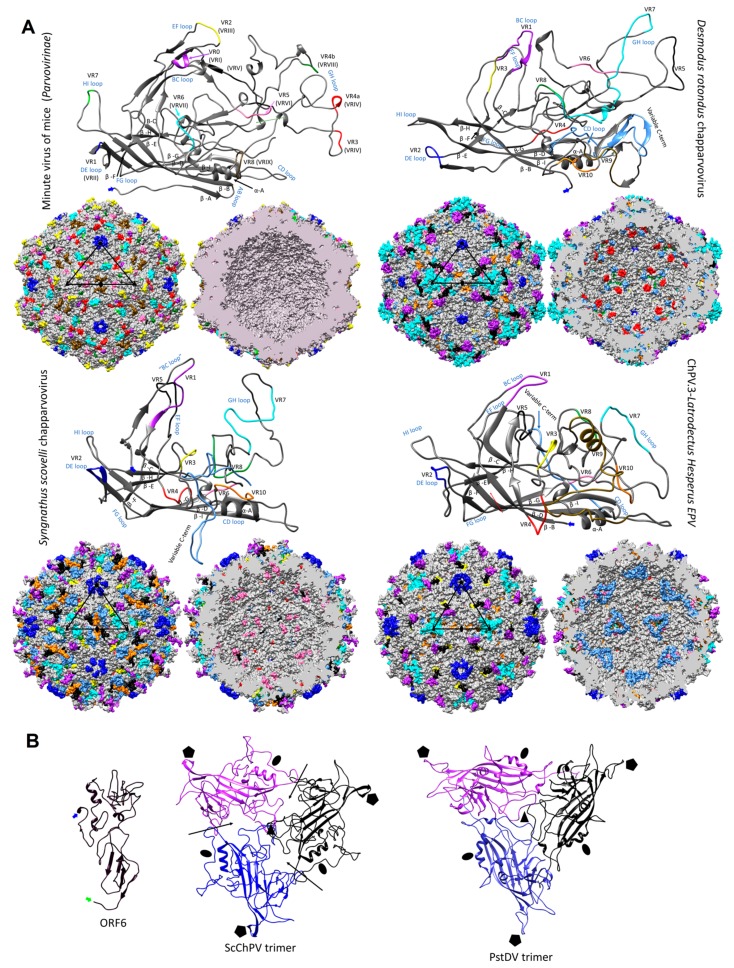
Figure 6.
Structural variation and assembly interfaces of chapparvoviruses (ChPVs). (A) Comparison of VP monomer ribbon diagrams of the protoparvovirus minute virus of mice (PDB ID: 1Z14) from subfamily Parvovirinae to homology models of an amniote, a fish, and a ChPV-derived EPV from an arthropod genome (ChPV.3-LatHes). Variable regions (VRs) of the same number are marked by the same color and mapped to the surface and luminal area of the T = 1 icosahedral capsid model constructed of 60 monomers. In the case of the minute virus of mice, the VRs are marked by both the traditional numbering established for dependoparvoviruses (Roman numerals) and by the special numbering applied for protoparvoviruses only (Arabic numerals). Blue signs indicate the names of the loops linking the beta strands of the conserved jelly roll core. Triangles mark the position of an asymmetric unit within the capsid, the five-fold symmetry axis is marked by a pentagon, the three-fold with the black filled triangles, and the two-fold with an ellipsoid. (B) Homology model of ORF6, the hypothetical structural protein of Syngnathus scovelli ChPV (ScChPV). The trimer of the ScChPV monomer model reveals a gap at each subunit interaction (arrows), unlike in the case of the trimer of even the hitherto smallest parvoviral capsid protein, Penaeus stylirostris densovirus. The gap might accommodate ORF6 in the assembled ScChPV capsid. Symmetry axes are marked by the same symbols as for panel A.