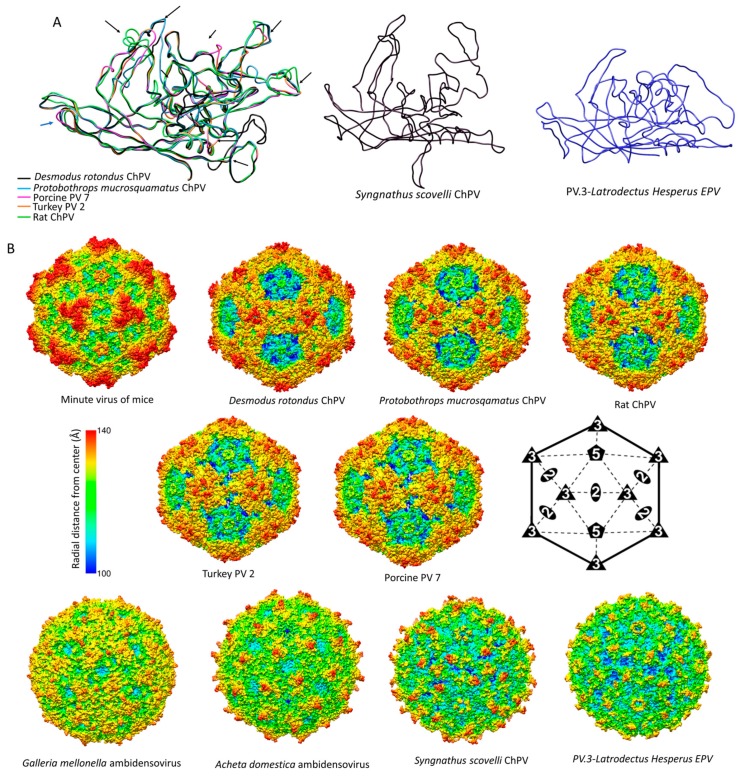
Figure 7.
Comparison of chapparvoviruses (ChPV) capsid models of various host affiliations. (A) Homology models, shown as ribbon diagrams, representing the probable three different ChPV structural protein types. The first panel shows superposition of VP monomer homology models of amniote ChPV capsids, including reptilian, avian, rodent, chiropteran, and ungulate representatives. Black arrows show variable regions (VRs) previously identified by aligning the VP protein sequences. The next two panels show homology models of capsid monomers from a fish ChPV and an endogenous chapparvoviral element from an arthropod genome. (B) Capsid surface morphology of amniote ChPV homology models compared to that of the polymer structure of a prototypic parvovirus, the minute virus of mice (MVM) (PDB ID: 1Z14 at 3.25 Å resolution). Capsids are orientated by their two-fold symmetry axes, as shown in the line diagram, and are radially colored. Below, the comparison of homology models of complete viral capsid surface morphology of the newly identified fish ChPV and arachnid endogenous chapparvoviral element is shown, with that of the actual capsid structure of two densoviruses (subfamily Densovirinae, genus Ambidensovirus) (PDB ID: 4MGU at 3.5 Å resolution for Acheta domestica densovirus and 1DNV at 3.7 Å for Galleria densovirus).