Fig. 4. Indirect and direct Merlin binding assays.
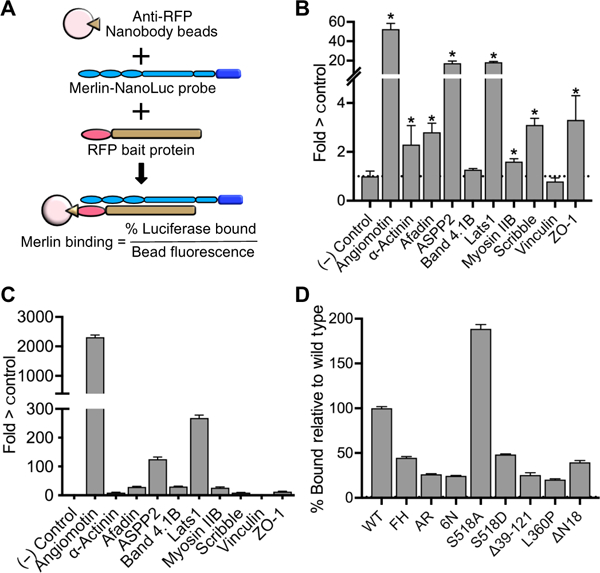
(A) Schematic diagram of the basic RFP pull-down assay with extracts of HEK 293T cells coexpressing RFP fused to bait proteins and a Merlin-Nanoluciferase probe (Merlin-NLuc) immunoprecipitated using a nanobody directed against RFP bound to magnetic beads. Data are expressed as % luciferase activity bound and normalized to red fluorescence on the bead (excitation 565 nm, emission 590). (B) Indirect interaction assays showing the relative luciferase activity that coimmunoprecipitated with the indicated RFP-tagged bait proteins from HEK 293T cells coexpressing the bait and Merlin-NLuc. RFP alone (no bait) was a negative control, and RFP-Angiomotin was a positive control. The dotted line represents the luciferase activity in the RFP-only negative control. Data are means relative to control ± SD; n = 3 independent experiments. *P > 0.05 by Student’s t test relative to control. (C) Direct binding assays showing the luciferase activity bound to purified RFP bait proteins that had been incubated with purified Merlin-NLuc. The dotted line represents the luciferase activity in the RFP-only negative control. Representative data of n > 3 independent biological experiments relative to control. (D) Relative binding of ASPP2 to the Merlin mutants used in the proximity biotinylation experiments (Merlin-FH, Merlin-AR, and Merlin-6N), the phosphorylation-deficient mutant S518A, the phosphomemetic mutant S518A, patient-derived mutations Δ39–121 and L360P, and the 18–amino acid N-terminal deletion mutant AN18. The dotted line represents the luciferase activity with wild-type (WT) Merlin. Data are means relative to wild type ± SD; n = 3 biological replicates.
