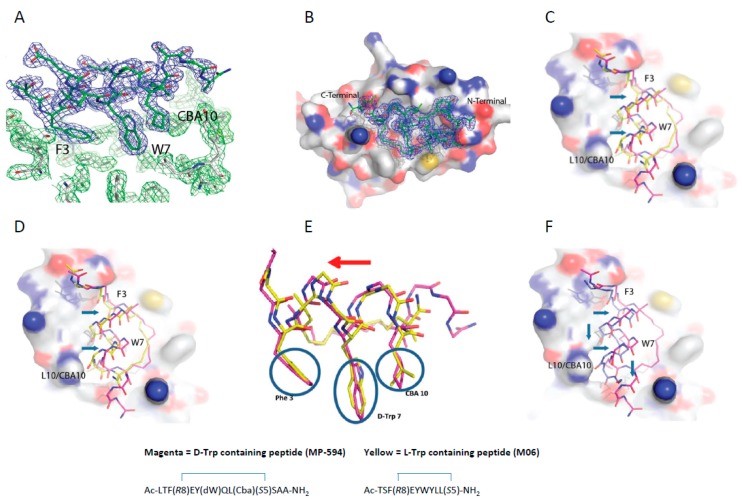
Figure 5.
(A) The three critical residues (F3, D-Trp7, and Cba10) responsible for the interaction of the peptide with Mdm2 project into a hydrophobic groove on the surface of Mdm2. 2Fo-Fc map for ATSP-7041 variant with a W7 to d-Trp substitution (MP-594) peptide is shown in blue and the 2Fo-Fc map for Mdm2 is shown in green (1.5σ). (B) Surface representation of Human Mdm2 (6–125) in complex with MP-594. The 2Fo-Fc electron density map for MP-594 is shown in blue mesh (1.5σ). (C) Alignment of MP-594 (magenta) with the M06 stapled peptide (chain C from PDB ID: 4UMN, yellow) (D) and the SAH-8 stapled peptide (chain B from PDB ID: 3V3B, cyan), both respectively in complex with Mdm2, highlighting the translation (blue arrows) of MP-594 across the peptide binding groove. (E) Overlay of MP-594 and M06, demonstrating that the d-Trp7 sidechain of MP-594 occupies the same volume of space as the Trp7 in M06 without perturbing the spatial positions of the other critical side chains in relation to each other or the overall α-helical fold of the constrained peptide. Only a global shift in register of the whole peptide occurs to maintain these critical interactions in their optimal positions with Mdm2. (F) Alignment of MP-594 (magenta) with the linear pMI N8A peptide (chain B from PDB ID: 3LNZ, blue), both in complex with Mdm2, where gross differences can be observed between both molecules in terms of displacement along and across the Mdm2 binding pocket (blue arrows).