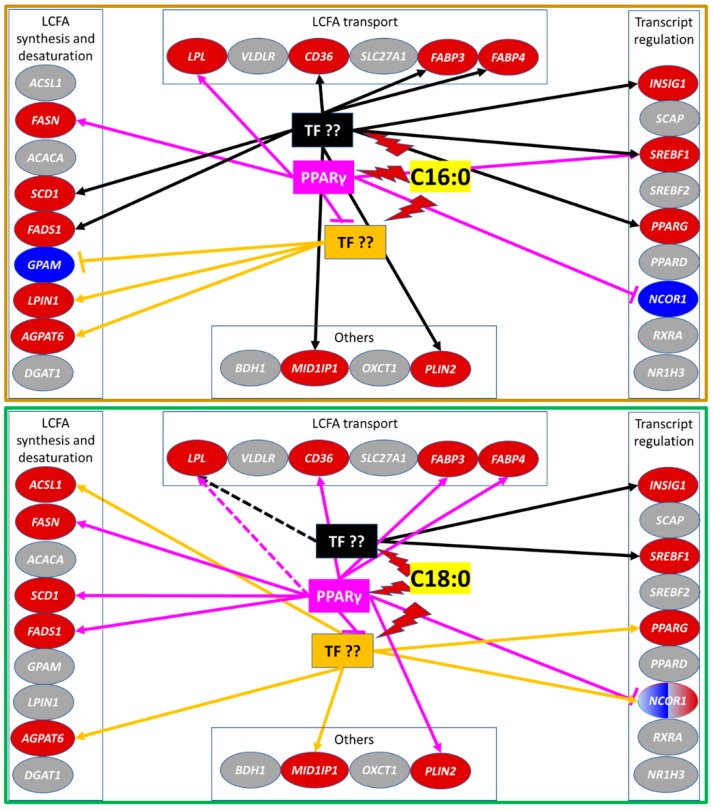
Figure 5.
Schematic summary of the results from the present experiment for the response of transcription of various genes to saturated LCFA C16:0 and C18:0 clustered in functional groups related to milk fat synthesis. It is also reported the possible interaction of LCFA with PPARγ or other unidentified transcription factors (TF) to elicit the observed responses. Grey shade denotes no significant change, red shade denotes upregulation, and blue shade denotes downregulation relative to control. In connecting lines, arrow head denotes induction while flat head denotes inhibition. Lightning symbol denotes induction of TF by the LCFA.Based on the results from our experiment, C16:0 controlled the expression of 4 genes (SREBF1, FASN, LPL, and NCOR1) via PPARγ or partly via this transcription factor (TF) (as was the case for SREBF1). The expression of other genes was controlled via other TFs. Data indicated that the activity of C16:0 was via one or more TF (in black) controlling the expression of the majority of the genes affected by C16:0 in our experiment that were independent from PPARγ (including SREBF1, which expression was only downregulated by the inhibition of PPARγ but was still strongly upregulated compared to control). Expression of several genes, such as GPAM, LPIN1, and AGPAT6, was controlled by C16:0 via a TF which activity was somewhat inhibited by the activity of PPARγ (in orange). C18:0 controlled the expression of 9 genes via PPARγ, including LPL which effect is only numerical and only partly via this TF. The expression of other genes was controlled via other TFs. Data indicated that the activity of C18:0 was via one or more TF (in black) independent from PPARγ controlling the expression of only 2 measured genes (INSIG1 and SREBF1). Expression of several genes (ACSL1, AGPAT6, MID1IP1, PPARG, and NCOR1) was upregulated by C18:0 via a TF which activity was inhibited by the activity of PPARγ (in orange).