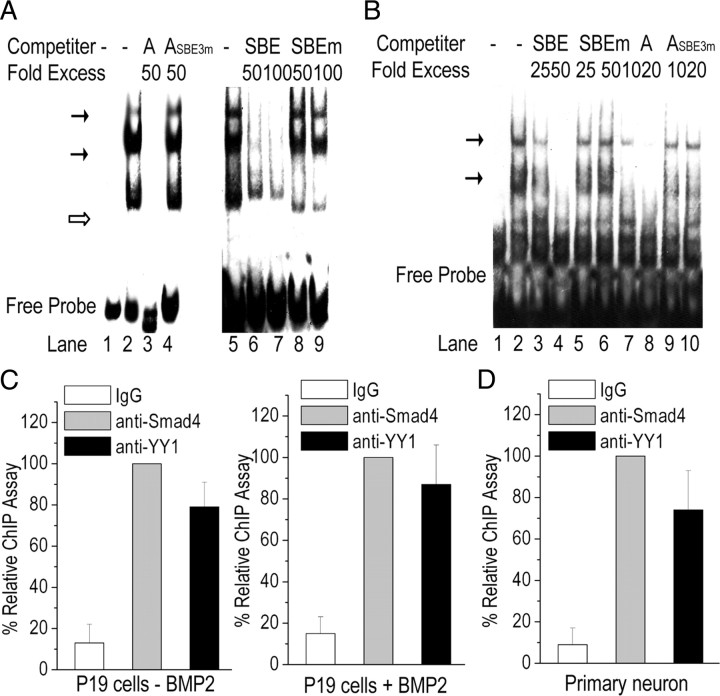
Figure 8.
Traditional EMSA and ChIP localize Smad4 and YY1 bindings to gat1 gene promoter. A, EMSA competition experiments. Nuclear protein was isolated from primary cortical neurons, and EMSA was performed using biotin-labeled oligonucleotide from gat1 gene promoter from −333 to −288 as a probe. Lane 1, free probe; lanes 2 and 5, no competitor; lanes 3 and 4, unlabeled wild-type probe (A) or mutant probe (ASBE3m) as competitor; lanes 6–9, unlabeled wild-type or mutant SBE oligonucleotides as competitor. Indicated are the specific complexes (solid arrow) or nonspecific complexes (open arrows). Results are representative of at least three independent experiments. B, EMSA competition experiments using biotin-labeled SBE oligonucleotides as a probe. Lane 1, free probe; lane 2, no competitor; lanes 3 and 4, wild-type SBE oligonucleotides as competitors; lanes 5 and 6, mutant SBE oligonucleotides as competitors; lanes 7 and 8, wild-type A oligonucleotides as competitors; lanes 9 and 10, mutant ASBE3m (with the functional SBE mutant) oligonucleotides as competitors. Results are representative of at least three independent experiments. C, D, Protein interactions at the region from −333 to −288 of mouse gat1 gene in P19 cells (C) or primary cortical neurons (D) were determined by ChIP assay. IgG was used as control. Chromatin immunoprecipitated DNA was analyzed by real-time PCR.