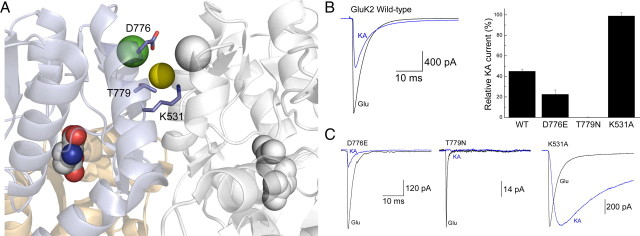
Figure 1.
Mutation of GluK2 dimer interface KA efficacy. A, Crystal structure of the dimeric GluK2 ligand-binding domain in complex with Glu and two cations (one green and one white sphere) and one anion (yellow sphere) (Protein Data Bank number 3G3F). B, Left, Representative response of wild-type (WT) GluK2 receptors to Glu (10 mm; black trace) and KA (1 mm; blue trace; patch number 030724p2). Right, Plot summarizing the effect of point mutations on the relative responsiveness of GluK2 receptors to KA. C, Membrane currents elicited by Glu (10 mm; black traces) and KA (1 mm; blue traces) for GluK2 mutants D776E (left; patch number 100121p6), T779N (middle; patch number 100128p2), and K531A (right; patch number 070215p2).