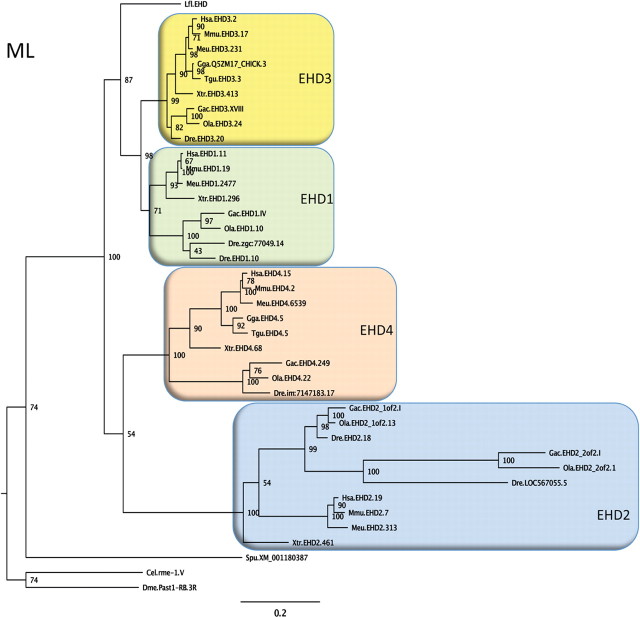
Figure 1.
Phylogeny of EHD proteins. A phylogenetic quartet-puzzling maximum likelihood tree using protein sequences is shown. The first three letters of the sequence names are abbreviations of the species names, then follows EHD subtype (according to the ENSEMBL or NCBI databases), and number for the chromosome, group, or scaffold on which the gene is located. A few sequences have been assigned administrative names in the databases and these have been retained in the tree. Bootstrap values are shown at the nodes. Species abbreviations: Cel, Caenorhabditis elegans; Dme, Drosophila melanogaster; Dre, Danio rerio (zebrafish); Gac, Gasterosteus aculeatus (three-spined stickleback); Gga, Gallus gallus; Hsa, Homo sapiens; Meu, Macropus eugenii (Tammar wallaby); Mmu, Mus musculus; Lfl, Lampetra fluviatilis (=l-EHD); Ola, Oryzias latipes (medaka); Spu, Strongylocentrotus purpuratus (California purple sea urchin); Tgu, Taeniopygia guttata (zebra finch); Xtr, Xenopus tropicalis (Western clawed frog). The tree suggests that a single invertebrate EHD gene was quadrupled before the radiation of jawed vertebrates, but detailed analyses of the chromosomal locations of the EHD genes across these species will be required before this conclusion can be firmly drawn. Accession codes, the alignment, and the neighbor-joining tree can be obtained upon request.