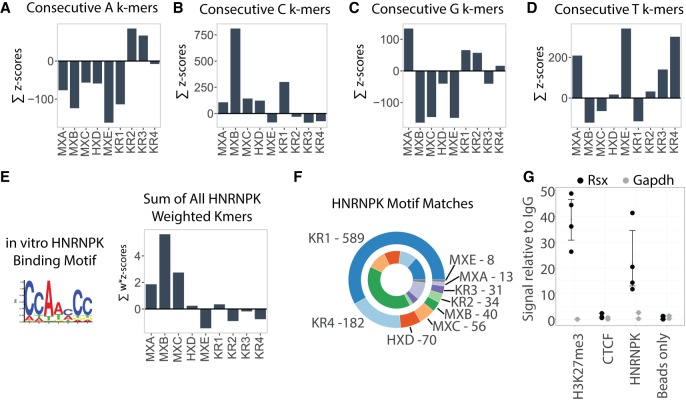
FIGURE 4.
Mononucleotide runs and HNRNPK-binding motifs enriched in specific Xist and Rsx repeats. (A–D) The sum of z-scores in each repeat for k-mers containing consecutive (A) A, (B) C, (C) G, and (D) T nucleotides. For this analysis we defined “consecutive” as at least two consecutive nucleotides of the specified identity and used k-mer length k = 5 (Materials and Methods). Repeat abbreviations as in Figure 2. (E) The sum of z-scores for all k-mers in each Xist and Rsx repeat domain after weighting the k-mers by the likelihood with which they fit the consensus HNRNPK-binding motif. Motif logo that describes the consensus HNRNPK-binding motif obtained from Ray et al. (2013) is also shown. (F) Component arcs of outer circle indicate the proportion and number of HNRNPK-binding motif matches detected by FIMO (P < 0.01) in each Xist and Rsx repeat domain. Component arcs of inner circle indicate the proportion of motif matches in each repeat domain normalized for domain length. Repeat abbreviations as in Figure 2. (G) Rsx enrichment relative to IgG control after RNA IP-qPCR in cultured fibroblasts from female M. domestica. For each antibody, left (black) is enrichment of Rsx, right (gray) is enrichment of Gapdh. The histone modification H3K27me3 is enriched on the inactive X in marsupials, so an association with Rsx was expected. CTCF has nanomolar affinity for RNA and along with “bead only”/no-antibody IP serves as a negative control demonstrating IP specificity (Kung et al. 2015). Dots represent values from replicate RNA IP experiments; error bars represent bootstrap 95% CI.