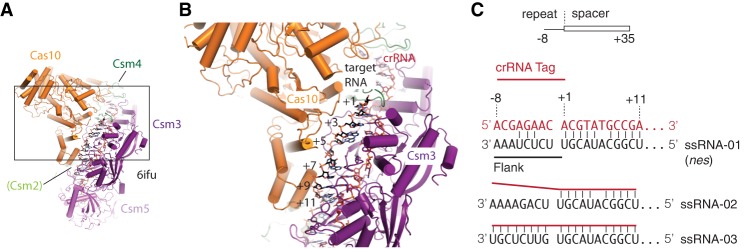
FIGURE 2.
The molecular environment of the crRNA–target RNA duplex. (A) An overview of the high-resolution cryo-EM structure of S. thermophilus Cas10-Csm bound to target RNA (6ifu). (B) The boxed region from A is magnified, showing that the +1 to +11 target RNA positions occupy a distinct environment, directly interacting with Cas10. The distal base pairs of the target RNA contact Csm2. (C) The 5′ tag region of crRNAs, in type Type III systems, is an 8 nt sequence derived from the repeat region of the CRISPR array. Base pairs between the crRNA and a foreign RNA target began downstream from the 5′ tag and are numbered starting with +1. Base pairs in the −1 to −8 region indicate a “self” nucleic acid, and this complementarity blocks Cas10-Csm–mediated interference. Three target RNAs that differ in the 5′ tag–3′ flank region of the duplex were used in this study. Csm3 occludes base-pairing at the +6 position.