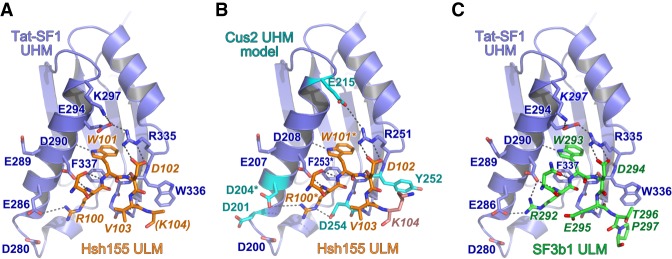
FIGURE 4.
Conserved interfaces revealed by Tat-SF1–ULM structures. (A) Hsh155 ULM (orange) bound to Tat-SF1 UHM (blue). The K104 side chain (parentheses) is disordered and lacks interpretable electron density. (B) Model of Hsh155 bound to Cus2 UHM. Interacting residues that differ from the Tat-SF1 homolog were substituted with a sterically compatible rotamer (cyan). Residues modified by mutagenesis (Cus2 D204 and F253; Hsh155 R100 and W101) are marked by asterisks. (C) Comparison of human SF3b1 ULM bound to Tat-SF1 UHM (PDB ID 6N3E).