Figure 36.
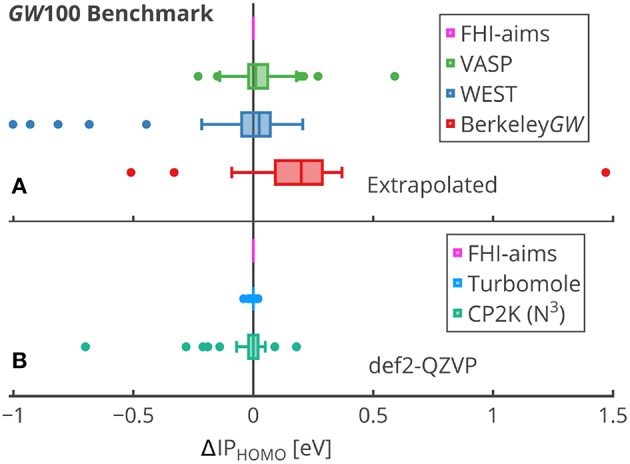
GW100 benchmark comparing IPHOMO energies computed at the G0W0@PBE level. FHI-aims is set as reference: ΔIPHOMO = IPHOMO(FHI-aims)−IPHOMO(X). (A) Comparison of extrapolated/converged results for VASP (Maggio et al., 2017), WEST (Govoni and Galli, 2018), BerkeleyGW (van Setten et al., 2015). Shown are the results from full-frequency treatments and iterative solutions of the QP equation. (B) Comparison of localized basis set codes using the Gaussian basis set def2-QZVP (Weigend and Ahlrichs, 2005) for Turbomole (no-RI) (van Setten et al., 2015) and the N3 implementation in CP2K (Wilhelm et al., 2018). Note that BN, O3, MgO, BeO, and CuCN are excluded for WEST, VASP and CP2K and that the BerkeleyGW and Turbomole data contain only a subset of 19 and 70 molecules, respectively. Box plot: Outliers represented by dots; boxes indicate the “interquartile range” measuring where the bulk of the data are.
