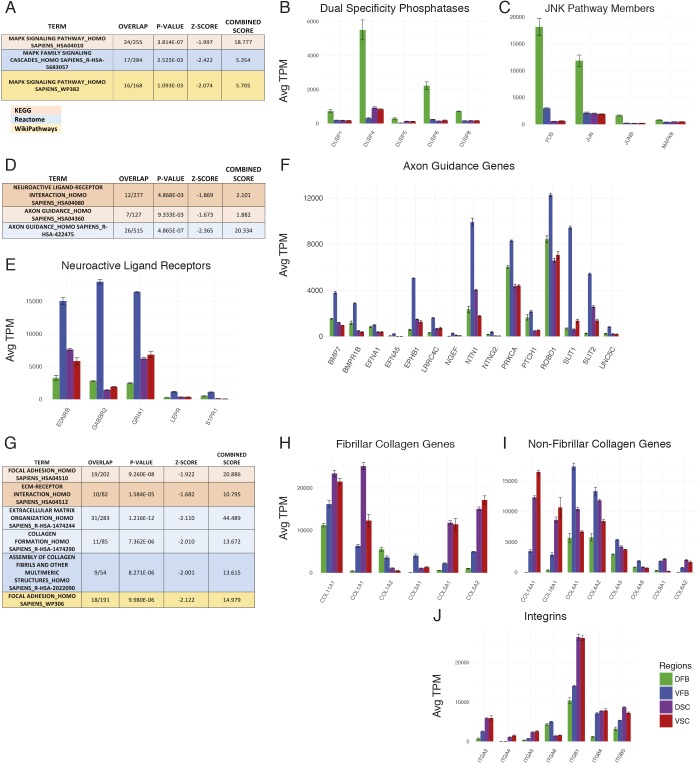
Fig. 3.
RNA-seq pathway analysis. (A) Analysis of the differentially utilized pathways in the DFB group; boxes show the MAPK pathway found in all analyses. (B) Expression of dual specificity phosphatase (DUSP) genes. (C) Expression of genes in the JNK/Jun pathway. (D) Analysis of the differentially utilized pathways in the VFB group; boxes show the axon guidance pathway and related neuroactive ligand receptor pathway found in KEGG and Reactome analyses. (E) Expression of selected genes in the neuroligand receptor pathway (KEGG HSA04080). (F) Expression of selected genes in the axon guidance pathway (KEGG HSA04360). (G) Analysis of the differentially utilized pathways in the spinal cord group; boxes show pathways involving extracellular matrix (ECM) function in all analyses. (H,I) Expression of fibrillary (H) and non-fibrillar (I) collagen genes. (J) Expression of integrin receptor genes. All gene expression values are average TPM (n=3 biological replicates of ESC-derived astrocytes for each group). Error bars represent s.d. of TPM.