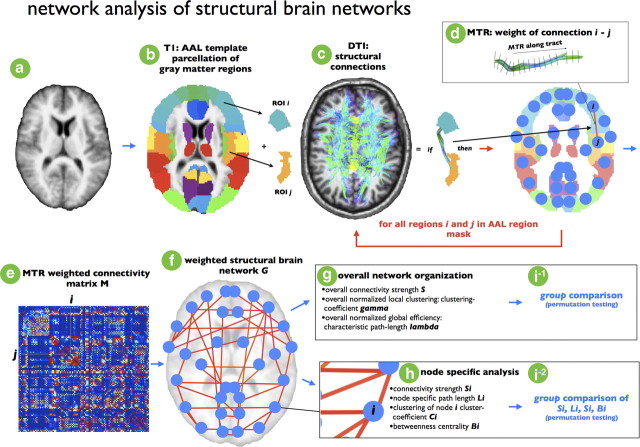
Figure 2.
Schematic overview of formation of individual structural brain networks. For each individual dataset, a structural brain network was constructed. First, for each dataset (a) from the AAL template consisting of 108 unique brain regions (54 each hemisphere), two regions, i and j, were selected (b). From the total collection of reconstructed white matter tracts (based on the DTI data), the white matter fibers interconnecting region i and region j were selected by selecting those tracts that touched both region i and region j. If this selection procedure revealed fibers between i and j (c), node i and node j in the brain network were interconnected by a connection. Furthermore, the weight wij of the connection between node i and j was selected as the MTR value along the selected fibers, reflecting the level of myelination of the selected tract (d), and wij was included in cell cij of the weighted connectivity matrix M. If no tract was found between region i and j, no connection was included in graph G, resulting in cij value of 0 in M. These steps were repeated for all regions i and j in the AAL template (e), resulting in a filled weighted connectivity matrix M, representing a weighted undirected graph G (f). Next, from the resulting brain network overall organizational characteristics (g) and node-specific organizational characteristics (h) (see supplemental materials and Fig. 1) were computed and compared between patients and healthy controls (i).