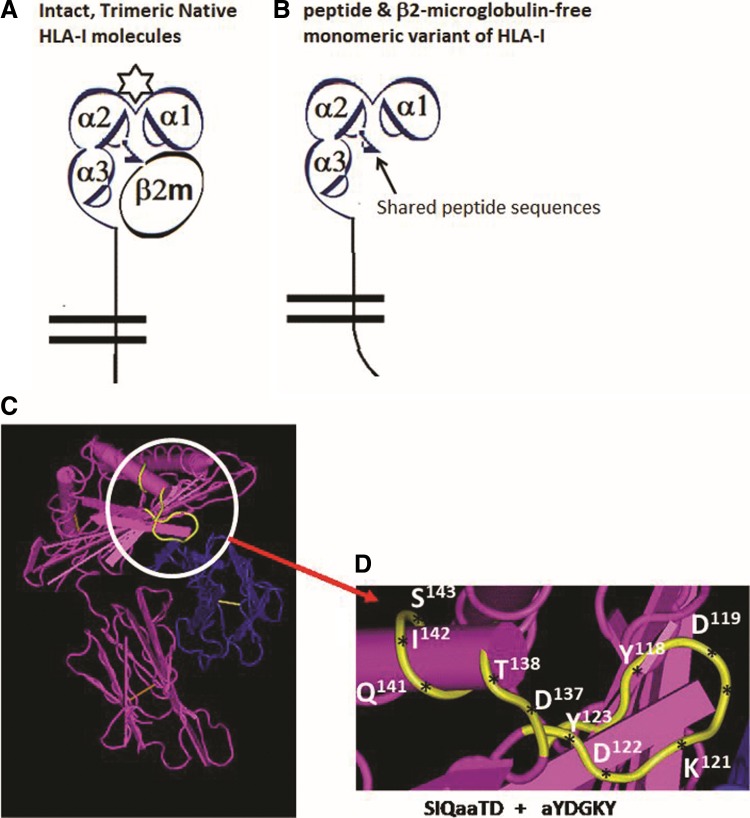
FIG. 2.
Structural variants of HLA class-I molecules. (A) Diagrammatic representation of an intact, native, and trimeric HLA-I molecules with α1, α2, and α3 helices of the heavy chain dimerized with β2m with the presence of a peptide (star) in the grove. The n-terminal end is extended into a bilayered lipid membrane. (B) Same as (A) but without β2m and peptide, called a monomeric variant, also known as open conformer. (C) Structure of an intact HLA without peptide but with β2m shown in blue. The amino acid shown in yellow is cryptic in the presence of β2m but gets exposed for immune recognition without β2m. The amino acid sequence in the yellow regions is shared by almost all HLA class-I molecules (Table 2). The monospecific mAb TFL-033 does not bind to the shared amino acid sequences. (D) The exact location, position, and configuration of the amino acid sequence of the shared epitopes, 115QFAYDGKDY123 and 137DTAAQI,142 recognized by the polyreactive mAb MEM-E/02 and all the anti-HLA-E mAbs included in group 8 category of mAbs are listed in Table 7. The antibody epitope prediction formulae(62) support the contention that the epitope for MEM-E/02 and other MEM series is a discontinuous sequence emerging from six to seven amino acid sequences separated by a long peptide sequence of the heavy chain of HLA-E molecule.