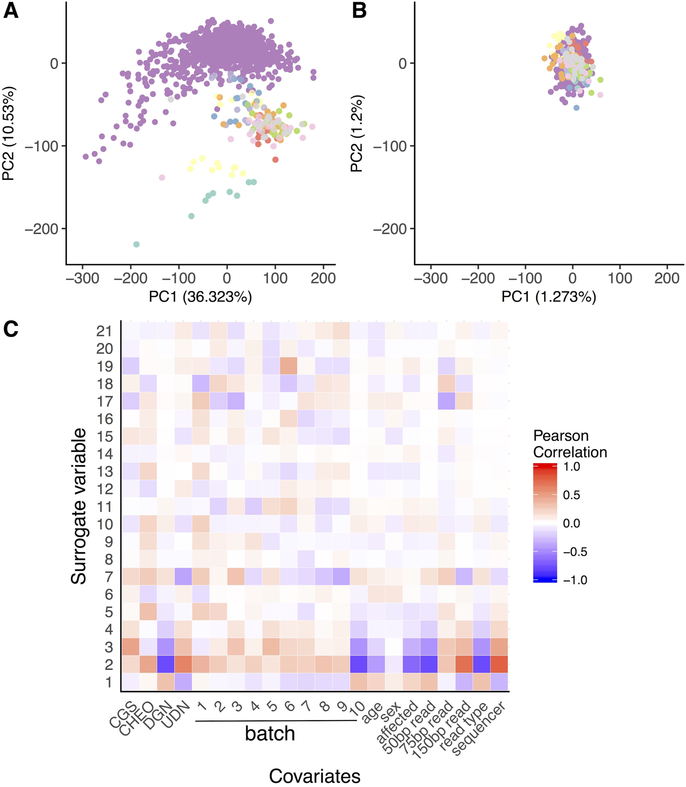
Extended Data Fig. 2. Correction for batch effects: Expression data.
Analyses performed on n = 909 DGN samples and 143 rare diseases (cases and family controls). a, Plot of first two principal components run on uncorrected gene expression data. Samples are coloured by batch. Largest cluster (green dots) are DGN control samples (n = 909). b, Plot of first two principal components run on gene expression data after regressing out significant surrogate variables found by SVA. c, Correlation between known covariates and all significant surrogate variables (SVs). We observed that SV2 is highly correlated with the read type, and the sequencing technology corresponding to differences between DGN and the other samples.