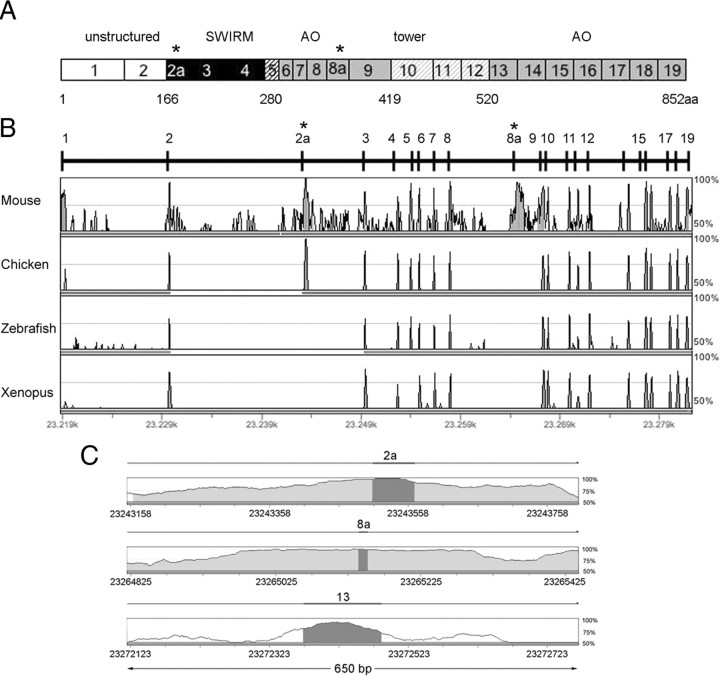
Figure 1.
Genomic organization of human LSD1 gene. A, Schematic representation of the human LSD1 protein domains together with its exons ranging from 1 to 19; asterisks indicate the location of annotated alternative exons (E2a and E8a). Different colors in grayscale indicate functional domains. N-terminal unstructured region (white) coded by exons 1–2, SWIRM domain (black) coded by exons 2–4, the SWIRM-oxidase connector (black striped) coded by exon 5, the amine oxidase domain (gray), coded by exons 6–9 and exons 13–19, and the tower domain (gray striped) coded by exons 10–12. Residue Met1 of this sequence corresponds to the first amino acid of the protein characterized (Shi et al., 2004). B, Human AOF2 alignment across vertebrates by GenomeVista browser (http://pipeline.lbl.gov/cgi-bin/GenomeVista). The “peaks and valleys” graphs represent percentage conservation at a given genomic coordinate between aligned sequences and the human sequence. Human exons are numbered. The top and bottom percentage bounds are shown to the right of every row. Regions of high conservation are colored as exons (dark gray) or noncoding (light gray). Conserved regions are defined as regions with identity of 70% or higher that are wider than or equal to “minimal conservation width” (100 bp). C, Enlarged view of 650 bp of the alignment between human and mouse intronic regions containing the two alternatively spliced exons (E2a and E8a) and one constitutively included exon (E13). Highlighted bars above the conservation area correspond to annotated E2a, E8a, and E13 of human AOF2. Dark gray areas within the conservation graph mark exons; light gray areas mark conserved (above 70%) nonexonic sequences.