Figure 8.
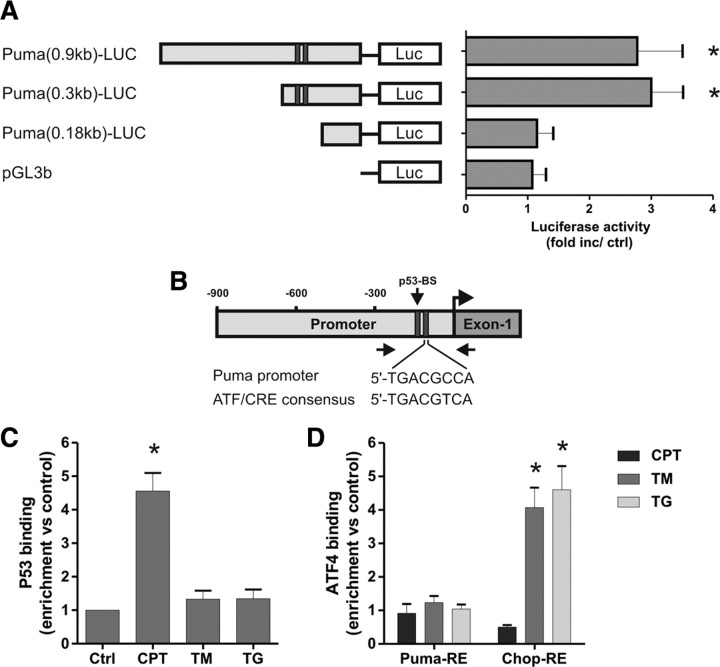
ATF4 does not directly activate the Puma promoter during ER stress. A, Neurons were cotransfected with the pGL3b or Puma–LUC reporter constructs and either pcDNA3 or pcDNA3–ATF4 and, after 4 h, treated with 200 nm thapsigargin. Luciferase activity was measured after 24 h and is reported as the ratio of luciferase activity produced in cells transfected with pcDNA3–ATF4 relative to empty pcDNA3 vector for each reporter construct (n = 4; *p < 0.05). B, Schematic of the Puma promoter showing the location of the putative ATF/CRE and p53 binding sites. Arrows indicate the approximate positions of the PCR primers used in the ChIP assays. C, Cortical neurons were treated with camptothecin (10 μm; CPT), tunicamycin (3 μg/ml; TM), or thapsigargin (1 μm; TG), and p53 binding to the Puma promoter was assessed after 8 h by ChIP assay. The level of p53 binding was quantified by real-time PCR and is reported as fold enrichment over untreated controls (Ctrl) (n = 5; *p < 0.01). D, Cortical neurons were treated as above, and binding of ATF4 was assessed by ChIP assay and real-time PCR using primers specifically targeting the putative ATF4 response elements (RE) in the Puma and Chop promoters. Data are reported as fold increase over untreated control samples for each promoter region (n = 5; *p < 0.01).