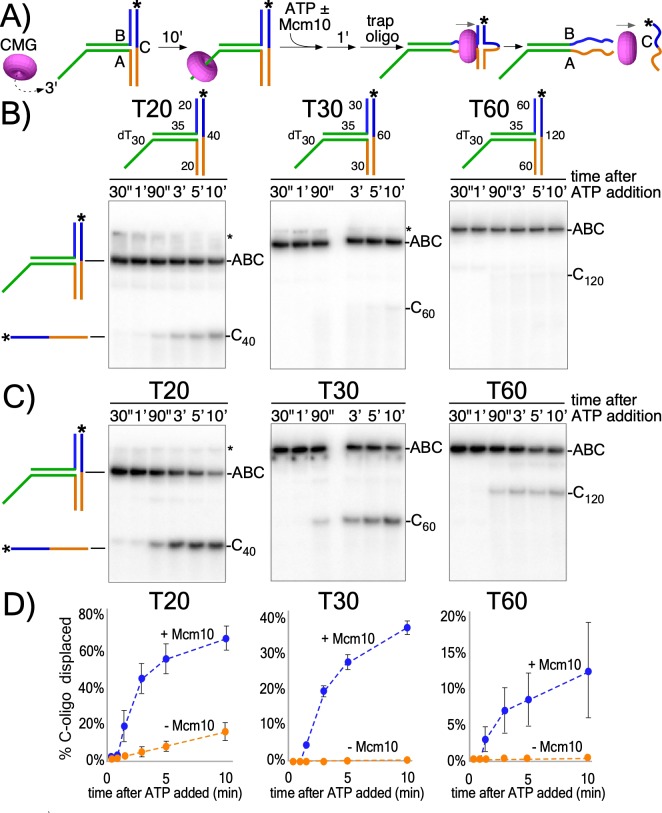
Figure 2. CMG translocates with force while encircling duplex DNA.
(A) Schematic of reactions using T-substrates with a 3’ dT30 ssDNA tail for CMG loading, a flush ss/ds junction and 35 bp of dsDNA (green) preceding the non-homologous arm blocks (blue and orange). See Figure 2—figure supplement 1 for details on the substrates. Unwinding the 32P-cross-bar oligo (labeled with a *) requires CMG to track with force while encircling dsDNA. (B) Native PAGE gel of time course reactions using CMG (no Mcm10) on T-substrates with different length arms. (C) Repeat of (B) with addition of Mcm10. (D) Plots of the data from (B) and (C). Values shown are the average of three independent experiments and the error bars show one standard deviation. The asterisks to the right of the gels indicate gel-shift of the substrate by CMG and Mcm10. See also Figure 2—figure supplements 2–3.
Figure 2—figure supplement 1. The T-shaped substrates shown in schematic form.
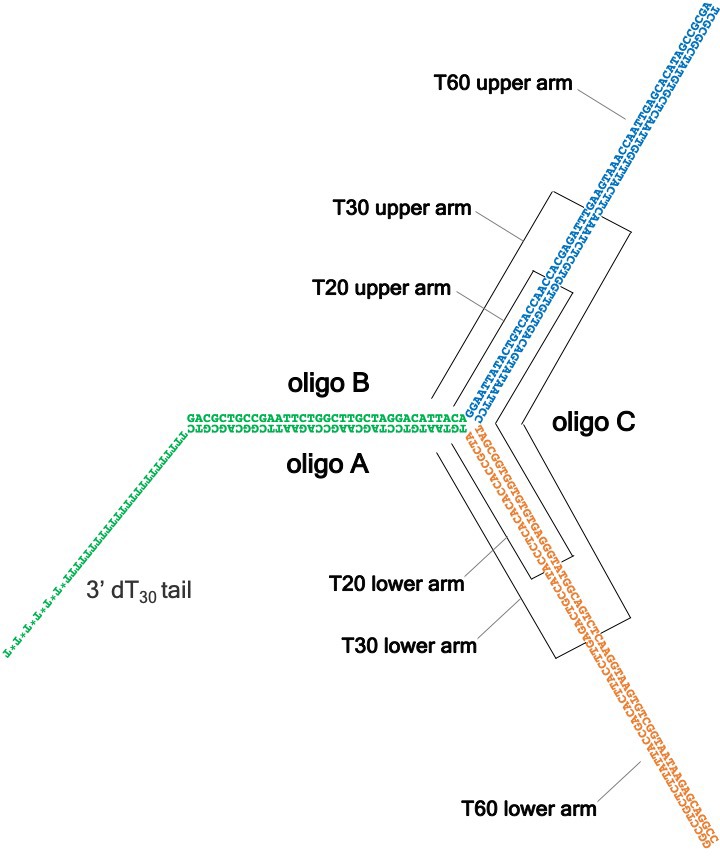
Figure 2—figure supplement 2. The 3’ dT30 tail is required for CMG loading onto the T-substrates.
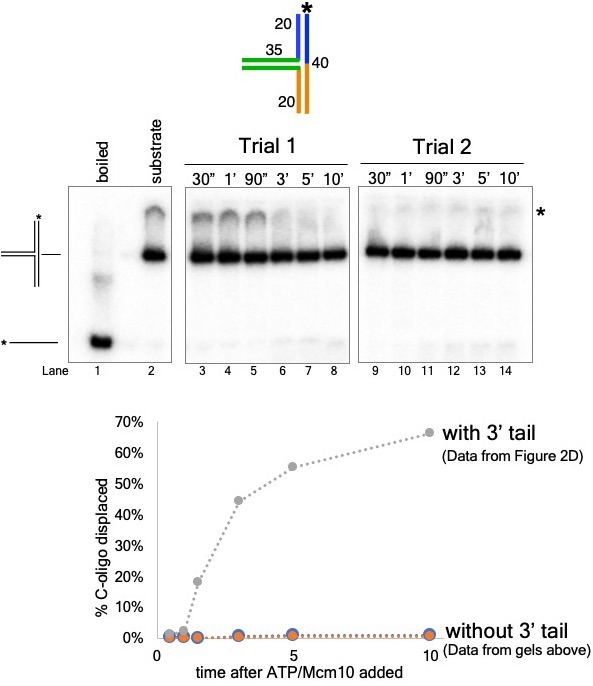
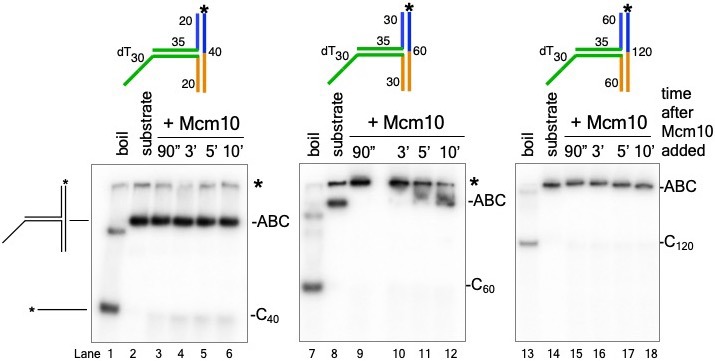
Figure 2—figure supplement 3. Mcm10 does not unwind the T-substrates without CMG.