Figure 3. CMG+Mcm10 strand interactions during duplex translocation.
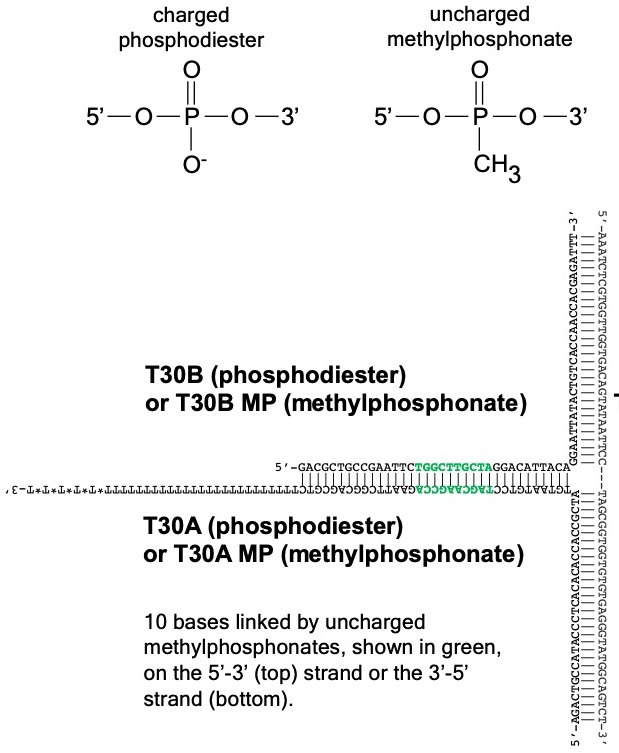
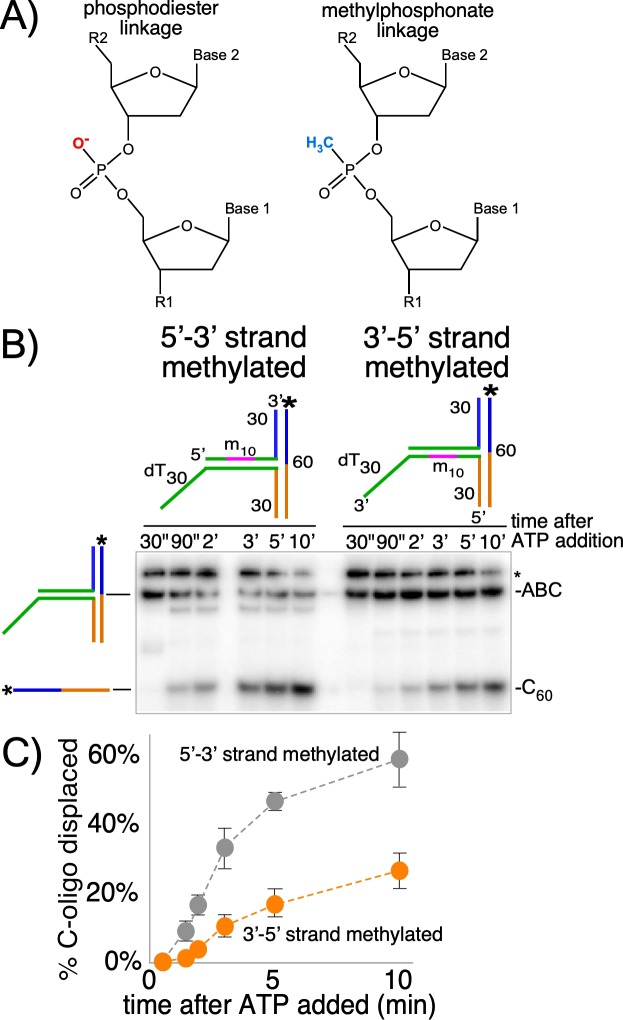
(A) Diagram of the naturally occurring negatively charged phosphodiester linkage in the phosphate backbone of a DNA chain (left) and the uncharged methylphosphonate linkages used in these experiments (right). (B) The experiment from Figure 2C (CMG+Mcm10) was repeated using a T30 substrate with 10 neutral methylphosphonate linkages (shown in pink in the schematics above the gel) in the 5’-3’ strand (left) or in the 3’-5’ strand (right) of the duplex. See Figure 3—figure supplement 2 and Supplementary file 1 for further details on the substrate. The * next to the gels indicates gel-shift of the substrate by CMG+Mcm10. (C) A plot of the data from (B) shows the averages of three independent trials using these substrates. The error bars show the standard deviations. Also see Figure 3—figure supplement 1.
Figure 3—figure supplement 1. Duplex strand ATPases contact only one strand while encircling duplex DNA.
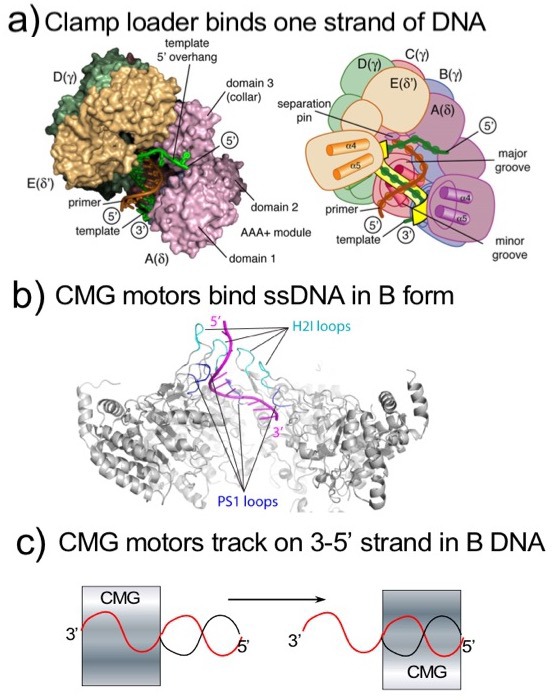
Figure 3—figure supplement 2. Design of substrates with methylphosphonate linkages in Figure 3.