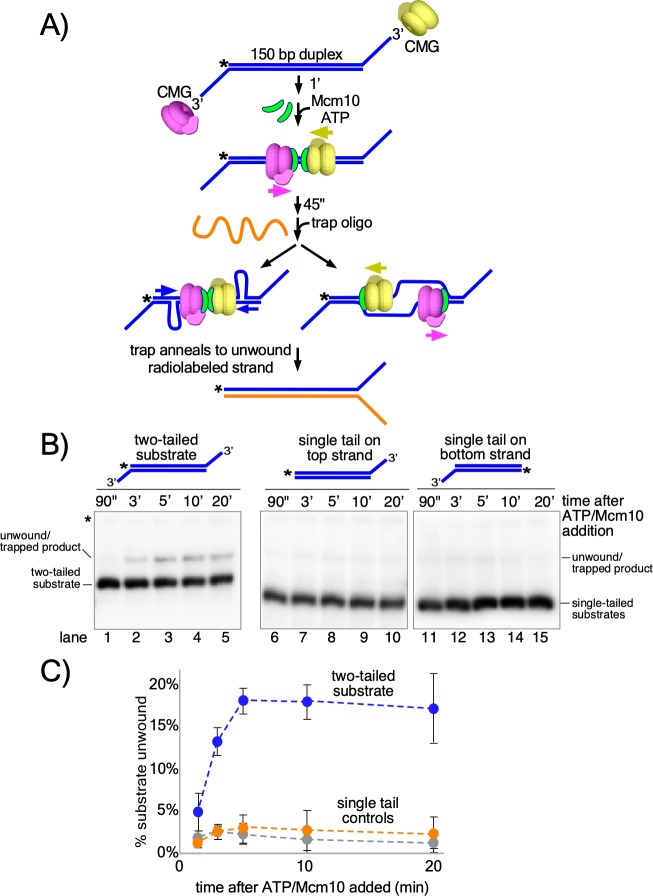
Figure 4. Head-to-head CMGs unwind dsDNA.
(A) Scheme of the reaction with the double tailed substrate, and two possible processes that could result in unwinding the DNA. The color scheme is the same as in Figure 1. CMG is mixed with the substrate on ice and then incubated at 30° C in the absence of ATP (top) to allow the reaction to reach temperature. 1’ later, ATP is added to allow CMGs to load onto the duplex in opposite orientations and block each other’s progress (middle). At the same time as ATP, Mcm10 is added to initiate the duplex unwinding reaction. 45’ later, an ssDNA trap oligo (orange) is added that quenches further CMG loading (Figure 4—figure supplement 1) and also anneals to the unwound radiolabeled product, creating a forked structure (bottom) that migrates at a distinct position in the gel from the substrate. Unwinding may occur either by: strand shearing (left arrow), or by strand expulsion (right arrow) to form CMGs that encircle ssDNA for conventional helicase activity. (B) Native PAGE gel time course of results using CMG + Mcm10. Lanes 1–5 show the reaction using the two-tailed substrate described in (A) while lanes 6–10 and 11–15 show control reactions using the same duplex with only a single tail at one end or the other. The migration of the substrates and unwound/trapped product are indicated to the left and right of the gel. (C) A plot of the data from (B) shows the averages of three independent trials using these substrates. The error bars show the standard deviations. See also Figure 4—figure supplements 2–3.
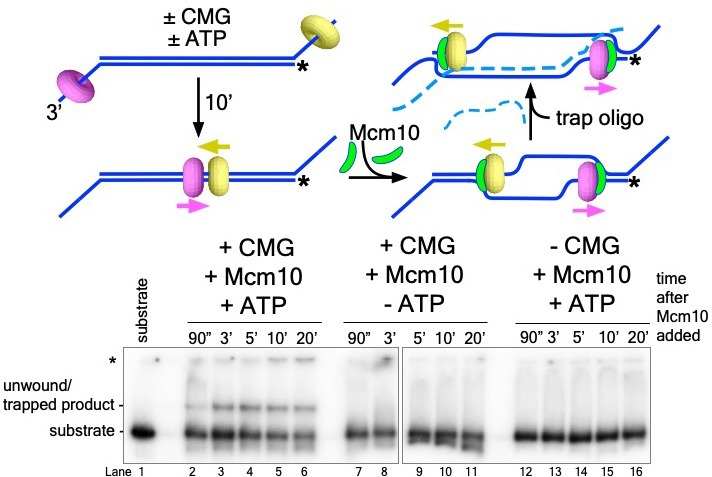
Figure 4—figure supplement 1. Addition of oligo trap prevents further loading of CMGM onto the two-tailed substrate.
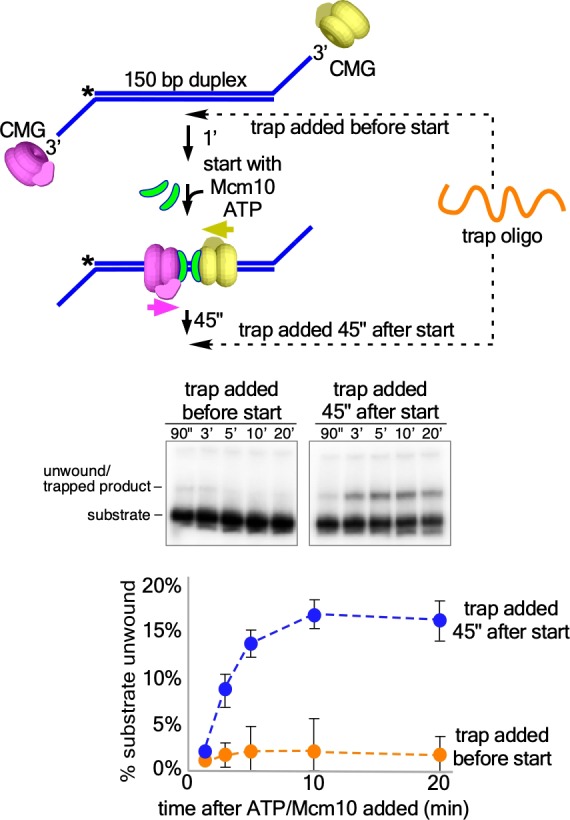
Figure 4—figure supplement 2. CMG alone does not unwind the origin-duplex substrate.
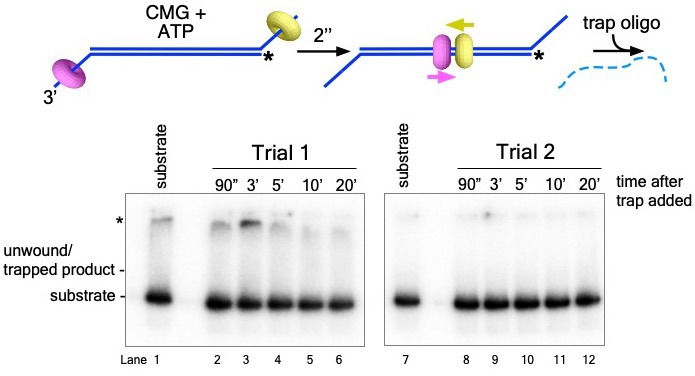
Figure 4—figure supplement 3. Unwinding of the origin-duplex substrate in Figure 4 requires ATP and CMG.