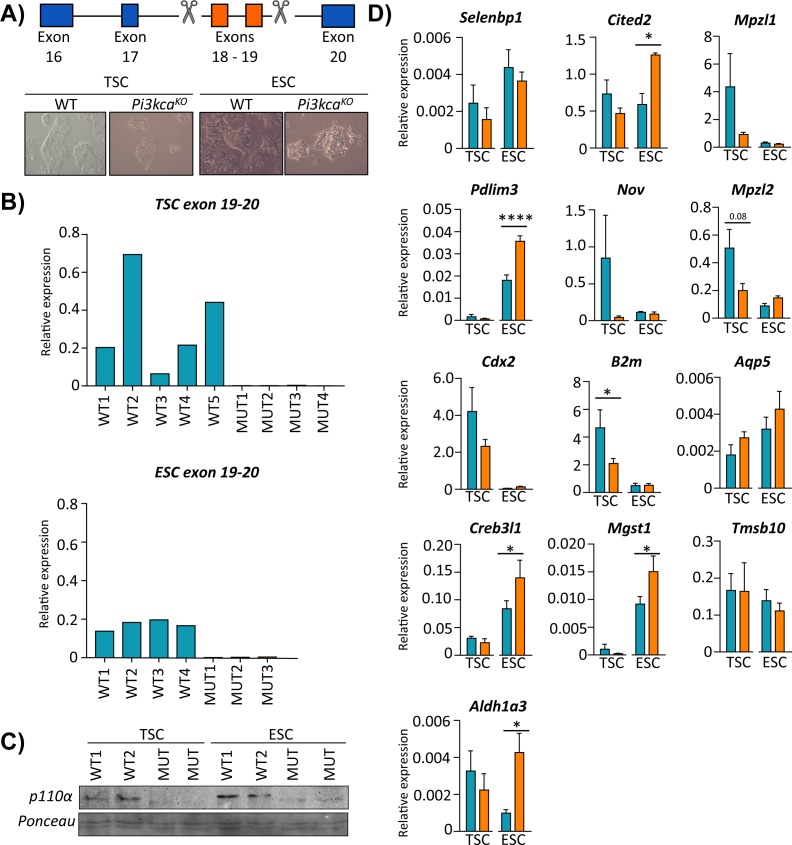
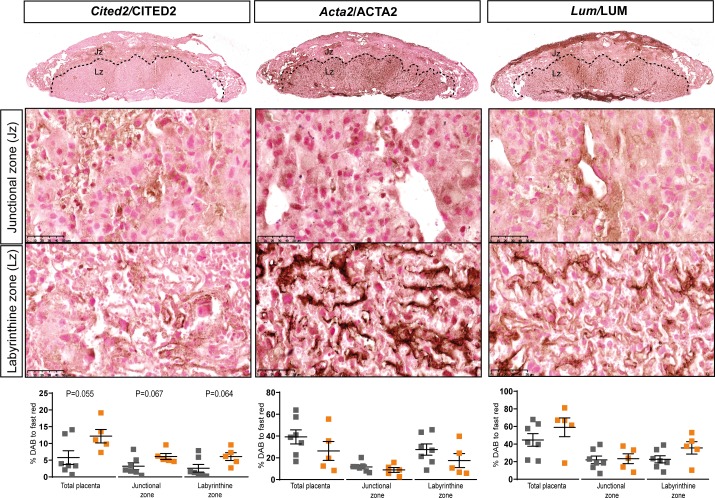
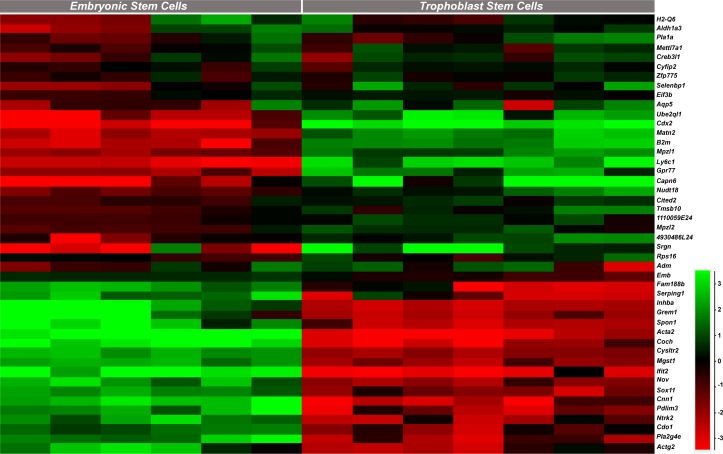
Figure 7. PI3K p110α differentially affects the expression of genes in the trophoblast and fetal cell lineages.
(A) Details of CRISPR/Cas9 design to delete exons 18–19 of Pik3ca in trophoblast or embryonic stem cells (TSC and ESC, respectively). The deletion in TSC (trophoblast stem cells) and ESC (embryonic stem cells) was confirmed by (B) qRT-PCR analysis of exons 18–19 of Pik3ca and (C) by Western blotting for p110α. (D) The expression of candidate genes in Pi3kca wildtype (WT) and mutant (Mut) TSC (n = 5 and n = 4, respectively) and ESC (n = 4 and n = 3, respectively). Data in (D) are normalized against housekeeping Sdha. *p<0.05, ****p<0.0001, two-ways ANOVA analysis followed by Fisher post hoc test. Data presented as means ± SEM.