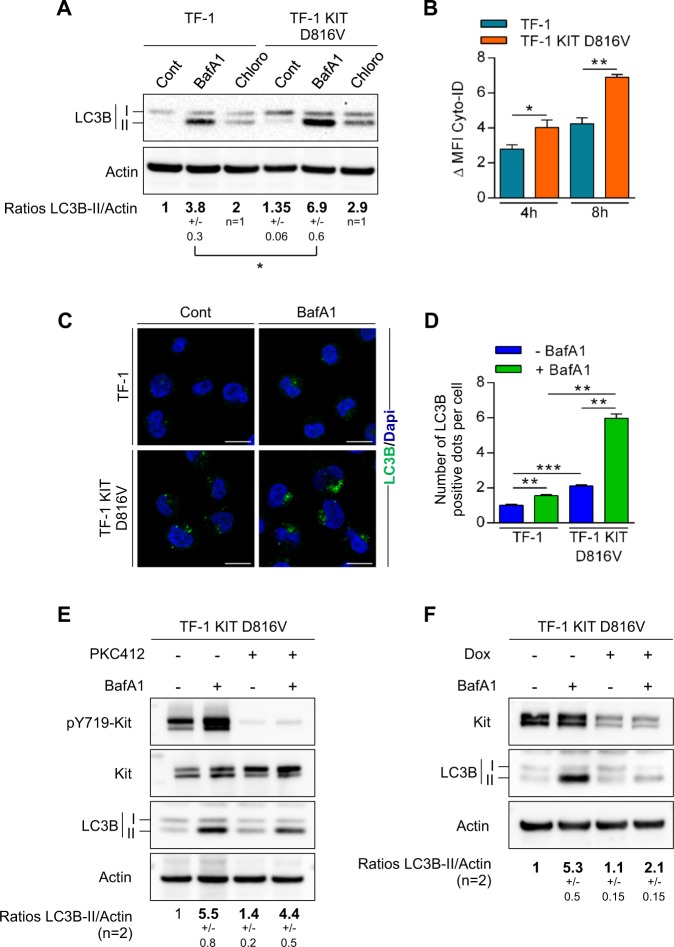
Fig. 1. KITD816V mutation increases autophagic flux in AML cells.
a–d Oncogenic KITD816V drives autophagy. a TF-1 or TF-1 KITD816V cells were incubated for 4 h with PBS, Bafilomycin A1 (20 nM, n = 3 ± SEM), or chloroquine (20 µM, n = 2 ± SD), and were then analyzed by immunoblotting using the appropriate antibodies. Numbers represent the LC3B-II/actin ratios obtained by densitometric analysis. b Cells were incubated for 4 or 8 h with vehicle or 20 µM of chloroquine, and then incubated with Cyto-ID before flow cytometry (n = 3 ± SEM). ΔMFI = MFI(chloroquine) − MFI(vehicle). c Cells were treated with vehicle or BafA1 for 2 h before LC3 staining, fluorescent labeling, and immunofluorescence analysis. d Quantification of LC3-positive autophagosomes (n = 3 ± SEM). e, f Pharmacological inhibition or genetic invalidation of a decrease in KIT autophagic flux. e TF-1 KITD816V cells were treated overnight with PKC412 at 1 µM; 20 nM of BafA1 was added at 2 h before cell lysis and western blotting. Numbers represent the LC3B-II/actin ratios obtained by densitometric analysis (n = 2 ± SD). f KITD816V cells were incubated for 3 days with doxycycline to induce expression of shRNA against KIT. Cells were treated for 2 h with BafA1 (20 nM) and then analyzed by western blotting. Numbers represent the LC3B-II/actin ratios obtained by densitometric analysis (n = 2 ± SD)